
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,528,011 – 21,528,153 |
| Length | 142 |
| Max. P | 0.636962 |

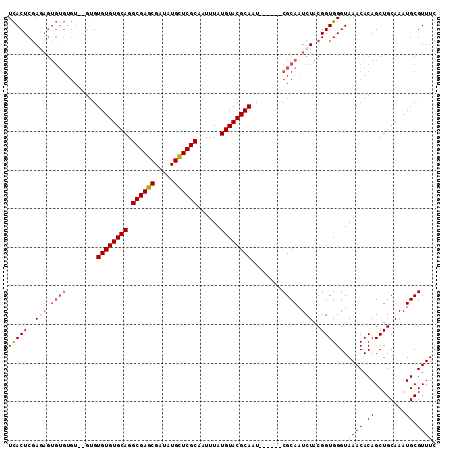
| Location | 21,528,011 – 21,528,117 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 86.75 |
| Mean single sequence MFE | -31.82 |
| Consensus MFE | -24.92 |
| Energy contribution | -24.42 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21528011 106 + 23771897 UCGGUAUCUGAAACUUUGGGCUAUCUUAGGAUAAUCCUUUCACUCGAUAGUGUGUGU--GUGUGUGUGCAGGCGAGCGAUAUGCUCGCAAUUUAUGUACGCAAU------CGCA .((.(((((((((....((..((((....))))..)))))))...)))).))..(((--(..((((((((.((((((.....))))))......))))))))..------)))) ( -34.70) >DroSec_CAF1 33407 108 + 1 UCGGAAUCUGAAACUUUGGGCUAUCUUAGGAUAAUCCUUUCACUCGAGAGUGUGUGUGUGUGUGUGUGCAGGCGAGCGAUAUGCUCGCAAUUUAUGUACGCAAU------CGCA ((((...))))......((..((((....))))..))...(((.(....).)))..((((..((((((((.((((((.....))))))......))))))))..------)))) ( -34.50) >DroSim_CAF1 33789 108 + 1 UCGGAAUCUGAAACUUUGGGCUAUCUUAGGAUAAUCCUUUCACUCAAGAGUGUGUGUGUGUGUGUGUGCAGGCGAGCGAUAUGCUCGCAAUUUAUGUACGCAAU------CGCA ((((...))))......((..((((....))))..))...((((....))))....((((..((((((((.((((((.....))))))......))))))))..------)))) ( -33.30) >DroYak_CAF1 8598 100 + 1 UCGGAAUCUGGAUCUUUGGACUAUCUUAGAAUAAUCCUUUUACUCGA--------------GUGUGUGCAGGCGAGCGAUAUGUUCGCAAUUUAUGUACGCAAUCGCAAUCGCA ..(((.(((((((.........))).))))....)))........((--------------.((((((((.((((((.....))))))......)))))))).))((....)). ( -24.80) >consensus UCGGAAUCUGAAACUUUGGGCUAUCUUAGGAUAAUCCUUUCACUCGAGAGUGUGUGU__GUGUGUGUGCAGGCGAGCGAUAUGCUCGCAAUUUAUGUACGCAAU______CGCA ........(((......(((.((((....)))).)))......)))................((((((((.((((((.....))))))......))))))))............ (-24.92 = -24.42 + -0.50)



| Location | 21,528,050 – 21,528,153 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 88.70 |
| Mean single sequence MFE | -32.77 |
| Consensus MFE | -26.64 |
| Energy contribution | -28.02 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597139 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21528050 103 + 23771897 UCACUCGAUAGUGUGUGU--GUGUGUGUGCAGGCGAGCGAUAUGCUCGCAAUUUAUGUACGCAAU------CGCAAUCUACGGUGGGUAAACACAGCUGCAAAUGCGUUUC ((((.((.(((..((((.--...((((((((.((((((.....))))))......))))))))..------))))..)))))))))..((((.((........)).)))). ( -33.80) >DroSec_CAF1 33446 105 + 1 UCACUCGAGAGUGUGUGUGUGUGUGUGUGCAGGCGAGCGAUAUGCUCGCAAUUUAUGUACGCAAU------CGCAAUCUACGGUGGGUAAACACAGCUGCAAAUGCGUUUC ((((.(((((.((((..((((((..((.....((((((.....))))))....))..))))))..------)))).))).))))))..((((.((........)).)))). ( -35.20) >DroSim_CAF1 33828 105 + 1 UCACUCAAGAGUGUGUGUGUGUGUGUGUGCAGGCGAGCGAUAUGCUCGCAAUUUAUGUACGCAAU------CGCAAUCUACGGUGGGUAAACACAGCUGCAAAUGCGUUUC ..(((((...(((..((((....((((((((.((((((.....))))))......))))))))..------))))...)))..)))))((((.((........)).)))). ( -33.90) >DroYak_CAF1 8637 97 + 1 UUACUCGA--------------GUGUGUGCAGGCGAGCGAUAUGUUCGCAAUUUAUGUACGCAAUCGCAAUCGCAAUCUACGGUGGGUAAACACAGCUGCAAAUGCGUUUC ((((((((--------------.((((((((.((((((.....))))))......)))))))).))((....))..........)))))).....((.......))..... ( -28.20) >consensus UCACUCGAGAGUGUGUGU__GUGUGUGUGCAGGCGAGCGAUAUGCUCGCAAUUUAUGUACGCAAU______CGCAAUCUACGGUGGGUAAACACAGCUGCAAAUGCGUUUC (((((..(((.((((........((((((((.((((((.....))))))......))))))))........)))).)))..)))))..((((.((........)).)))). (-26.64 = -28.02 + 1.38)


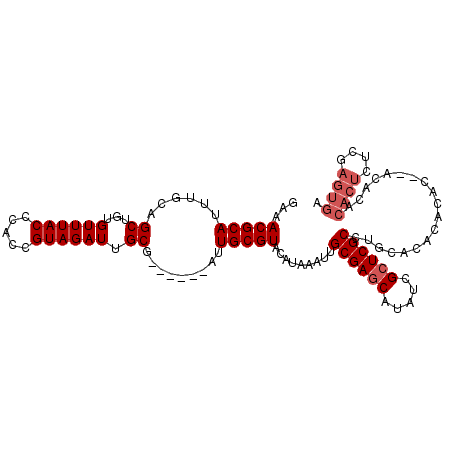
| Location | 21,528,050 – 21,528,153 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 88.70 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -19.56 |
| Energy contribution | -20.81 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.600103 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21528050 103 - 23771897 GAAACGCAUUUGCAGCUGUGUUUACCCACCGUAGAUUGCG------AUUGCGUACAUAAAUUGCGAGCAUAUCGCUCGCCUGCACACACAC--ACACACACUAUCGAGUGA ...(((((.(((((((((((......)))...)).)))))------).))))).........((((((.....))))))............--.....((((....)))). ( -28.60) >DroSec_CAF1 33446 105 - 1 GAAACGCAUUUGCAGCUGUGUUUACCCACCGUAGAUUGCG------AUUGCGUACAUAAAUUGCGAGCAUAUCGCUCGCCUGCACACACACACACACACACUCUCGAGUGA ...(((((.(((((((((((......)))...)).)))))------).))))).........((((((.....))))))...................((((....)))). ( -29.20) >DroSim_CAF1 33828 105 - 1 GAAACGCAUUUGCAGCUGUGUUUACCCACCGUAGAUUGCG------AUUGCGUACAUAAAUUGCGAGCAUAUCGCUCGCCUGCACACACACACACACACACUCUUGAGUGA ...(((((.(((((((((((......)))...)).)))))------).))))).........((((((.....))))))...................((((....)))). ( -29.20) >DroYak_CAF1 8637 97 - 1 GAAACGCAUUUGCAGCUGUGUUUACCCACCGUAGAUUGCGAUUGCGAUUGCGUACAUAAAUUGCGAACAUAUCGCUCGCCUGCACACAC--------------UCGAGUAA ...(((((.((((((.(((((((((.....))))).)))).)))))).)))))....................(((((..((....)).--------------.))))).. ( -24.30) >consensus GAAACGCAUUUGCAGCUGUGUUUACCCACCGUAGAUUGCG______AUUGCGUACAUAAAUUGCGAGCAUAUCGCUCGCCUGCACACACAC__ACACACACUCUCGAGUGA ...(((((......((...((((((.....)))))).)).........))))).........((((((.....))))))...................((((....)))). (-19.56 = -20.81 + 1.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:08:49 2006