| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,491,815 – 2,491,910 |
| Length | 95 |
| Max. P | 0.861532 |

| Location | 2,491,815 – 2,491,910 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 80.06 |
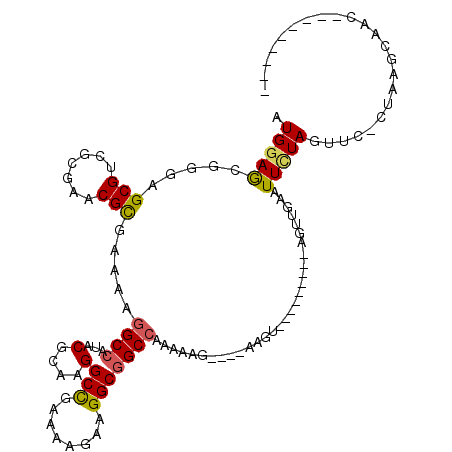
| Mean single sequence MFE | -24.85 |
| Consensus MFE | -17.71 |
| Energy contribution | -17.68 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
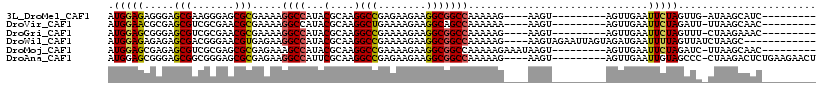
>3L_DroMel_CAF1 2491815 95 - 23771897 AUGGAGAGGGAGCGAAGGGAGCGCGAAAAGGCCAUACGCAAGGCCGAGAAGAAGGCGGCCAAAAAG----AAGU---------AGUUGAAUUCUAGUUG-AUAAGCAUC--------- .(((((.....(((.......))).....((((...(....)(((........)))))))......----....---------.......)))))(((.-...)))...--------- ( -22.30) >DroVir_CAF1 133026 95 - 1 AUGGAACGCGAGCGUCGCGAACGCGAAAAGGCCAUACGCAAGGCUGAAAAGAAGGCAGCCAAAAAA----AAGU---------AGUUGAAUUCUAGAUU-UUAAGCAAC--------- ......((((.....))))...((.....((((........))))....((((..((((.......----....---------.))))..)))).....-....))...--------- ( -20.10) >DroGri_CAF1 110411 95 - 1 AUGGAGCGGGAGCGUCGCGAACGCGAAAAGGCCAUACGCAAGGCCGAAAAGAAGGCGGCCAAAAAG----AAGU---------AGUUGAAUUCUAGUUU-CUAAGAAAC--------- ..(((((.((((..((((....))))...((((...(....)(((........)))))))......----....---------.......)))).))))-)........--------- ( -27.60) >DroWil_CAF1 152385 102 - 1 AUGGAGAGAGAGCGACGGGAACGUGAGAAGGCCAUACGCAAGGCCGAAAAGAAGGCGGCCAAAAAG----AAGUAGAAUUAGUAGAUGAAUUUUAGUUAUCUAAGC------------ ......(((.(((.(((....))).....((((...(....)(((........)))))))......----...(((((((........)))))))))).)))....------------ ( -24.20) >DroMoj_CAF1 120775 99 - 1 AUGGAGCGAGAGCGUCGCGAGCGCGAGAAAGCCAUACGCAAGGCCGAAAAGAAGGCGGCCAAAAAGAAAUAAGU---------AGUUGAAUUCUAGAUC-UUAAGCAAC--------- ((((.((....)).((((....)))).....))))..((..(((((.........))))).........((((.---------....(....).....)-))).))...--------- ( -24.60) >DroAna_CAF1 95449 104 - 1 AUGGAGCGGGAGCGGCGGGAGCGCGAGAAGGCCAUUCGCAAGGCCGAGAAGAAGGCGGCCAAAAAG----AAGU---------AGUUGAAUUGUAGCCC-CUAAGACUCUGAAGAACU ..((((((((.((.(((....))).....((((...(....)(((........)))))))......----....---------............))))-)...).))))........ ( -30.30) >consensus AUGGAGCGGGAGCGUCGCGAACGCGAAAAGGCCAUACGCAAGGCCGAAAAGAAGGCGGCCAAAAAG____AAGU_________AGUUGAAUUCUAGUUC_CUAAGCAAC_________ .(((((.....(((.......))).....((((...(....)(((........)))))))..............................)))))....................... (-17.71 = -17.68 + -0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:57 2006