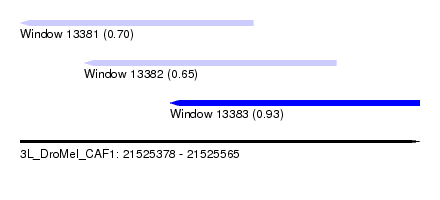
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,525,378 – 21,525,565 |
| Length | 187 |
| Max. P | 0.925846 |

| Location | 21,525,378 – 21,525,487 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 86.01 |
| Mean single sequence MFE | -36.38 |
| Consensus MFE | -26.75 |
| Energy contribution | -26.38 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700650 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21525378 109 - 23771897 CCAGGUCUGUCCCAGUCGGGGCACACUC-ACAUCUGGAUCCUUGCAACCAUGCAUCCCAUGCAUCCAUGCAUCCAUGUGCAUUUCUGGGUC--------UCUGGAUCUGCGGAUUUGC .((((((((((((....)))))......-......((((((.((((....)))).((((((((..((((....))))))))....))))..--------...))))))..))))))). ( -34.30) >DroSec_CAF1 30781 100 - 1 CCAGGUCUGUCCCAGUCGGGGCACACUC-GCAUCUGGAUCCUUGCAUCCAUGCAUCGCA---------GCAUCCAUGUGCAUUUCUGGGUC--------UCUGGAUCUGCGGAUUUGC .((((((((((((....))))))....(-((((((((((((.(((((..((((......---------))))....))))).....)))))--------)..)))..)))))))))). ( -33.90) >DroSim_CAF1 30853 100 - 1 CCAGGUCUGUCCCAGUCGGGGCACACUC-GCAUCUGGAUCCUUGCAUCCAUGCAUCCCA---------GCAUCCAUGUGCAUUUCUGGGUC--------UCUGGAUCUGCGGAUUUGC .((((((((((((....))))))....(-((((((((((((.(((((..((((......---------))))....))))).....)))))--------)..)))..)))))))))). ( -33.90) >DroYak_CAF1 5850 117 - 1 CCAGGUCUGUCCCAGUCGGGGUAUACUAGGCAUCUGGAUCCUUGCAACCAUGCAUCC-AGGCAUUCAGGCAUCCAUGUGCAUUUCUGGAUCUCUGGAUCUCUGGAUCUGCGGAGUUGC (((((((((((((....)))).....)))))..))))......(((((..(((((((-(((.(((((((.(((((.(......).))))).))))))).))))))..))))..))))) ( -43.40) >consensus CCAGGUCUGUCCCAGUCGGGGCACACUC_GCAUCUGGAUCCUUGCAACCAUGCAUCCCA_________GCAUCCAUGUGCAUUUCUGGGUC________UCUGGAUCUGCGGAUUUGC (((((((((((((....)))))..............(((((.((((....))))..............((((....))))......)))))...........))))))).)....... (-26.75 = -26.38 + -0.38)

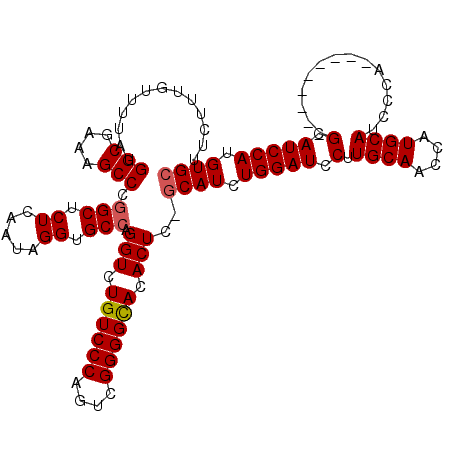
| Location | 21,525,408 – 21,525,526 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 90.46 |
| Mean single sequence MFE | -36.45 |
| Consensus MFE | -29.62 |
| Energy contribution | -29.93 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21525408 118 - 23771897 UUCUUUGUUUUAUGGCGAAAGCCCAGCUCUCAAUAGGUGCCAGGUCUGUCCCAGUCGGGGCACACUC-ACAUCUGGAUCCUUGCAACCAUGCAUCCCAUGCAUCCAUGCAUCCAUGUGC .............(((....)))..((.(.....(((((...(((.((((((....)))))).))).-.)))))(((....((((...((((((...))))))...)))))))..).)) ( -33.90) >DroSec_CAF1 30811 109 - 1 UUCUUUGUUUUAUGGCGAAAGCCCGGCUCUCAAUAGGUGCCAGGUCUGUCCCAGUCGGGGCACACUC-GCAUCUGGAUCCUUGCAUCCAUGCAUCGCA---------GCAUCCAUGUGC .............(((....))).(((.((.....)).))).(((.((((((....)))))).))).-((((.(((((.((((((....))))....)---------).))))).)))) ( -37.00) >DroSim_CAF1 30883 109 - 1 UUCUUUGUUUUAUGGCGAAAGCCCGGCUCUCAAUAGGUGCCAGGUCUGUCCCAGUCGGGGCACACUC-GCAUCUGGAUCCUUGCAUCCAUGCAUCCCA---------GCAUCCAUGUGC .............(((....))).(((.((.....)).))).(((.((((((....)))))).))).-((((.(((((.((((((....))))....)---------).))))).)))) ( -37.00) >DroYak_CAF1 5888 118 - 1 UUCUUUGUUUUAUGGCGAAAGCCCGGCUCUCAAUAGGUGCCAGGUCUGUCCCAGUCGGGGUAUACUAGGCAUCUGGAUCCUUGCAACCAUGCAUCC-AGGCAUUCAGGCAUCCAUGUGC .............(((....)))..((.(.....(((((((.(((.(..(((....)))..).))).)))))))((((.((((.....((((....-..)))).)))).))))..).)) ( -37.90) >consensus UUCUUUGUUUUAUGGCGAAAGCCCGGCUCUCAAUAGGUGCCAGGUCUGUCCCAGUCGGGGCACACUC_GCAUCUGGAUCCUUGCAACCAUGCAUCCCA_________GCAUCCAUGUGC .............(((....))).(((.((.....)).))).(((.((((((....)))))).)))..((((.(((((.(.((((....))))..............).))))).)))) (-29.62 = -29.93 + 0.31)

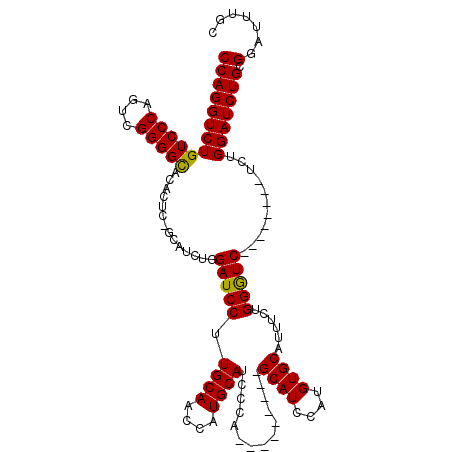
| Location | 21,525,448 – 21,525,565 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 97.45 |
| Mean single sequence MFE | -34.05 |
| Consensus MFE | -32.58 |
| Energy contribution | -32.45 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21525448 117 - 23771897 CCAUCUCCAUCAUAAACAACUGGCUCUUCUCGCCCAUAUUUCUUUGUUUUAUGGCGAAAGCCCAGCUCUCAAUAGGUGCCAGGUCUGUCCCAGUCGGGGCACACUC-ACAUCUGGAUC .....(((...........((((..(((.(((((.(((...........))))))))))).))))........(((((...(((.((((((....)))))).))).-.)))))))).. ( -31.70) >DroSec_CAF1 30842 117 - 1 CCAUCUCCAUCAUAAACAACUGGCUCUUCUCGCCCAUAUUUCUUUGUUUUAUGGCGAAAGCCCGGCUCUCAAUAGGUGCCAGGUCUGUCCCAGUCGGGGCACACUC-GCAUCUGGAUC .....(((...........((((..(((.(((((.(((...........))))))))))).))))........((((((..(((.((((((....)))))).))).-))))))))).. ( -34.40) >DroSim_CAF1 30914 117 - 1 CCAUCUCCAUCAUAAACAACUGGCUCUUCUCGCCCAUAUUUCUUUGUUUUAUGGCGAAAGCCCGGCUCUCAAUAGGUGCCAGGUCUGUCCCAGUCGGGGCACACUC-GCAUCUGGAUC .....(((...........((((..(((.(((((.(((...........))))))))))).))))........((((((..(((.((((((....)))))).))).-))))))))).. ( -34.40) >DroYak_CAF1 5927 118 - 1 CCAUCUCCAUCAUAAACAACUGGCUCUUCUCGCCCAUAUUUCUUUGUUUUAUGGCGAAAGCCCGGCUCUCAAUAGGUGCCAGGUCUGUCCCAGUCGGGGUAUACUAGGCAUCUGGAUC .....(((...........((((..(((.(((((.(((...........))))))))))).))))........(((((((.(((.(..(((....)))..).))).)))))))))).. ( -35.70) >consensus CCAUCUCCAUCAUAAACAACUGGCUCUUCUCGCCCAUAUUUCUUUGUUUUAUGGCGAAAGCCCGGCUCUCAAUAGGUGCCAGGUCUGUCCCAGUCGGGGCACACUC_GCAUCUGGAUC .....(((...........((((..(((.(((((.(((...........))))))))))).))))........((((((..(((.((((((....)))))).)))..))))))))).. (-32.58 = -32.45 + -0.13)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:08:43 2006