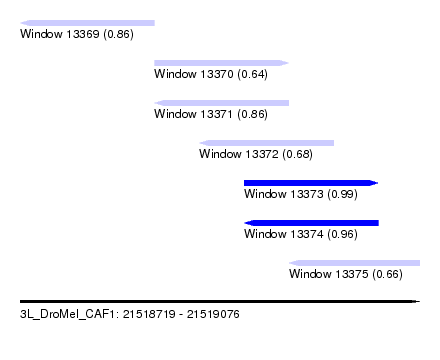
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,518,719 – 21,519,076 |
| Length | 357 |
| Max. P | 0.985194 |

| Location | 21,518,719 – 21,518,839 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -43.13 |
| Consensus MFE | -43.41 |
| Energy contribution | -42.97 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.83 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21518719 120 - 23771897 GAGUUCCCAGGCAGCGCGUUCGAGGCGGUAUGUAUCUUCGACUCACCCCCGACUCUAUUUAUCUCACGCGCCGUCCUGCUCUGCGUGUGCCGUCUCGCUCUGGCGCGCUCUCUUCGCUCU ((((....(((.((((((((((((((((((.........((.((......)).)).........((((((..(.....)..))))))))))))))))....)))))))).)))..)))). ( -41.80) >DroSec_CAF1 24019 120 - 1 GAGUUCCCAGGCAGCGCGUUCGAGGCGGUAUGUAUCUUCGACUCACCCCCGACUCUAUUUAUCUCGCGCGCCGUCCUGCUCUGCGCGUGCCGUCUCGCUCUGGCGCGCUCUCUUCGCUCU ((((....(((.(((((((((((((((((((........((.((......)).))..........(((((..(.....)..))))))))))))))))....)))))))).)))..)))). ( -43.80) >DroSim_CAF1 24133 120 - 1 GAGUUCCCAGGCAGCGCGUUCGAGGCGGUAUGUAUCUUCGACUCACCCCCGACUCUAUUUAUCUCGCGCGCCGUCCUGCUCUGCGCGUGCCGUCUCGCUCUGGCGCGCUCUCUUCGCUCU ((((....(((.(((((((((((((((((((........((.((......)).))..........(((((..(.....)..))))))))))))))))....)))))))).)))..)))). ( -43.80) >consensus GAGUUCCCAGGCAGCGCGUUCGAGGCGGUAUGUAUCUUCGACUCACCCCCGACUCUAUUUAUCUCGCGCGCCGUCCUGCUCUGCGCGUGCCGUCUCGCUCUGGCGCGCUCUCUUCGCUCU ((((....(((.(((((((((((((((((..........((.((......)).))..........((((((.(.......).)))))))))))))))....)))))))).)))..)))). (-43.41 = -42.97 + -0.44)

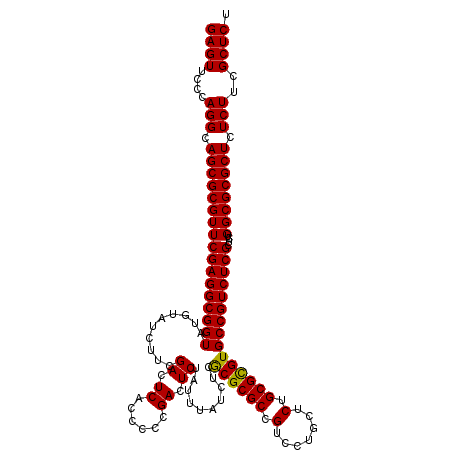
| Location | 21,518,839 – 21,518,959 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -43.50 |
| Consensus MFE | -41.03 |
| Energy contribution | -41.03 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21518839 120 + 23771897 UGCCAGCAUCCCCAAAAGCGAUAUGGUAUGUACAGCCCGAGCUGAAUGUGUAUUGGGGCUGUUCAGUUGUCGGUGACACGGGGGAUUGUAACUCACUCCCCCUUUUUGGGGCUUAUCCGA .........((((((((((((((.....((.(((((((.(((.......))..).))))))).))..))))).......((((((.((.....)).)))))))))))))))......... ( -43.30) >DroSec_CAF1 24139 119 + 1 UGCCAGCAUCCCCAAAAGCGAUAUGGUAUGUACAGCCCGAGCAGAAUGUGUAUUGGGGCUGUUCAGUUGUCGGUGACACGGGGGAUUGUAACUCACUCCCCCUU-UUGGGGCUUAUCCGA .........((((((((.(((((.....((.(((((((.((((.....)))..).))))))).))..))))).......((((((.((.....)).))))))))-))))))......... ( -43.60) >DroSim_CAF1 24253 119 + 1 UGCCAGCAUCCCCAAAAGCGAUAUGGUAUGUACAGCCCGAGCAGAAUGUGUAUUGGGGCUGUUCAGUUGUCGGUGACACGGGGGAUUGUAACUCACUCCCCCUU-UUGGGGCUUAUCCGA .........((((((((.(((((.....((.(((((((.((((.....)))..).))))))).))..))))).......((((((.((.....)).))))))))-))))))......... ( -43.60) >consensus UGCCAGCAUCCCCAAAAGCGAUAUGGUAUGUACAGCCCGAGCAGAAUGUGUAUUGGGGCUGUUCAGUUGUCGGUGACACGGGGGAUUGUAACUCACUCCCCCUU_UUGGGGCUUAUCCGA .........((((((...(((((.....((.(((((((.(((.......))..).))))))).))..))))).......((((((.((.....)).))))))...))))))......... (-41.03 = -41.03 + 0.00)

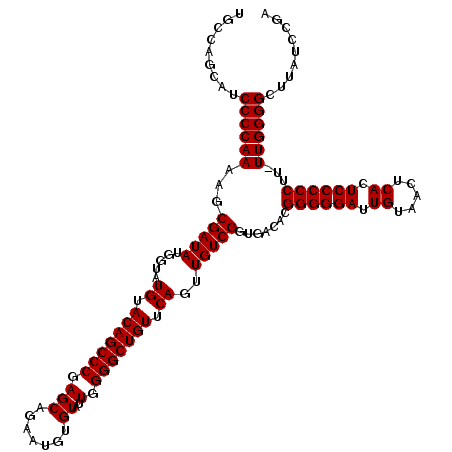
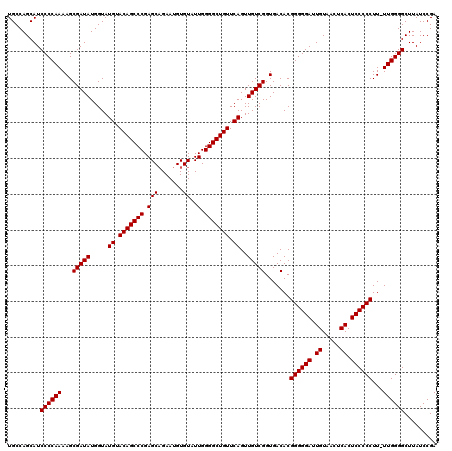
| Location | 21,518,839 – 21,518,959 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -40.33 |
| Consensus MFE | -38.10 |
| Energy contribution | -38.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21518839 120 - 23771897 UCGGAUAAGCCCCAAAAAGGGGGAGUGAGUUACAAUCCCCCGUGUCACCGACAACUGAACAGCCCCAAUACACAUUCAGCUCGGGCUGUACAUACCAUAUCGCUUUUGGGGAUGCUGGCA ((((.....((((((((.((((((.((.....)).)))))).((((...))))..((.(((((((.................))))))).))...........))))))))...)))).. ( -40.33) >DroSec_CAF1 24139 119 - 1 UCGGAUAAGCCCCAA-AAGGGGGAGUGAGUUACAAUCCCCCGUGUCACCGACAACUGAACAGCCCCAAUACACAUUCUGCUCGGGCUGUACAUACCAUAUCGCUUUUGGGGAUGCUGGCA ((((.....((((((-((((((((.((.....)).)))))).((((...))))..((.(((((((.................))))))).))...........))))))))...)))).. ( -40.33) >DroSim_CAF1 24253 119 - 1 UCGGAUAAGCCCCAA-AAGGGGGAGUGAGUUACAAUCCCCCGUGUCACCGACAACUGAACAGCCCCAAUACACAUUCUGCUCGGGCUGUACAUACCAUAUCGCUUUUGGGGAUGCUGGCA ((((.....((((((-((((((((.((.....)).)))))).((((...))))..((.(((((((.................))))))).))...........))))))))...)))).. ( -40.33) >consensus UCGGAUAAGCCCCAA_AAGGGGGAGUGAGUUACAAUCCCCCGUGUCACCGACAACUGAACAGCCCCAAUACACAUUCUGCUCGGGCUGUACAUACCAUAUCGCUUUUGGGGAUGCUGGCA ((((.....((((((...((((((.((.....)).)))))).((((...))))..((.(((((((.................))))))).)).............))))))...)))).. (-38.10 = -38.10 + -0.00)

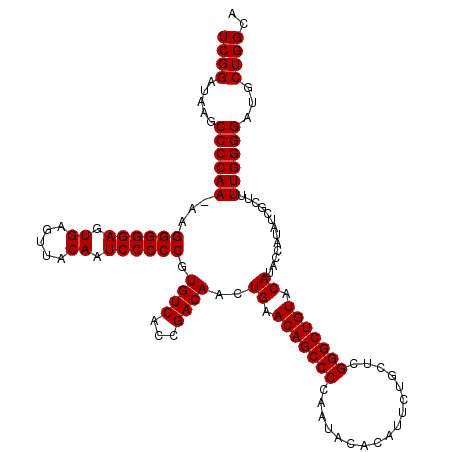
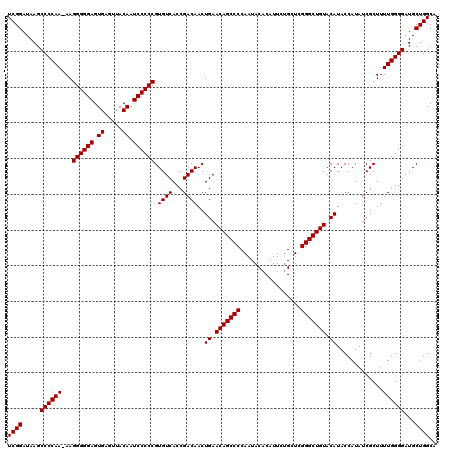
| Location | 21,518,879 – 21,518,999 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -35.47 |
| Consensus MFE | -35.47 |
| Energy contribution | -35.47 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.680648 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21518879 120 - 23771897 GAACUACCCCACUCGUUUGCCCGAGUGUAGAGGUACUGCUUCGGAUAAGCCCCAAAAAGGGGGAGUGAGUUACAAUCCCCCGUGUCACCGACAACUGAACAGCCCCAAUACACAUUCAGC (((...(..((((((......))))))..).((..(((.(((((..............((((((.((.....)).)))))).((((...)))).))))))))..))........)))... ( -35.60) >DroSec_CAF1 24179 119 - 1 GAAAUACCCCACUCGUUUGCCCGAGUGUAGAGGUACUGCUUCGGAUAAGCCCCAA-AAGGGGGAGUGAGUUACAAUCCCCCGUGUCACCGACAACUGAACAGCCCCAAUACACAUUCUGC (((...(..((((((......))))))..).((..(((.(((((...........-..((((((.((.....)).)))))).((((...)))).))))))))..))........)))... ( -35.40) >DroSim_CAF1 24293 119 - 1 GAAAUACCCCACUCGUUUGCCCGAGUGUAGAGGUACUGCUUCGGAUAAGCCCCAA-AAGGGGGAGUGAGUUACAAUCCCCCGUGUCACCGACAACUGAACAGCCCCAAUACACAUUCUGC (((...(..((((((......))))))..).((..(((.(((((...........-..((((((.((.....)).)))))).((((...)))).))))))))..))........)))... ( -35.40) >consensus GAAAUACCCCACUCGUUUGCCCGAGUGUAGAGGUACUGCUUCGGAUAAGCCCCAA_AAGGGGGAGUGAGUUACAAUCCCCCGUGUCACCGACAACUGAACAGCCCCAAUACACAUUCUGC (((...(..((((((......))))))..).((..(((.(((((..............((((((.((.....)).)))))).((((...)))).))))))))..))........)))... (-35.47 = -35.47 + -0.00)

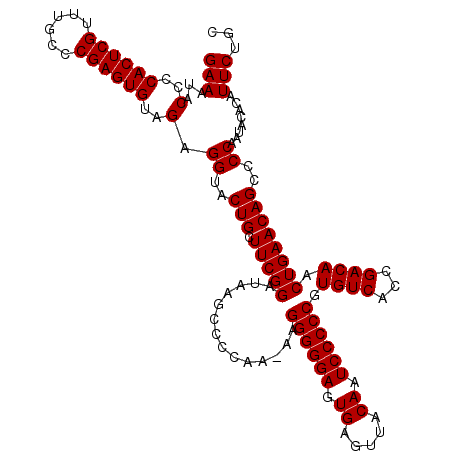
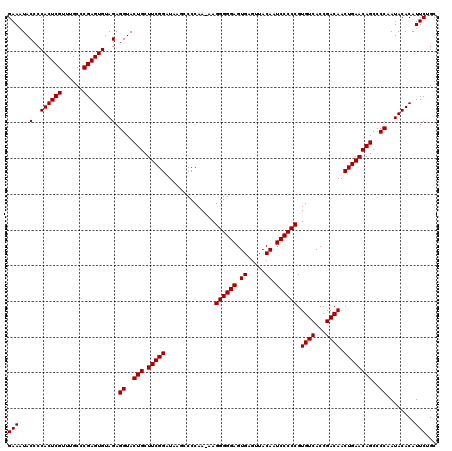
| Location | 21,518,919 – 21,519,039 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -41.30 |
| Consensus MFE | -39.77 |
| Energy contribution | -39.77 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21518919 120 + 23771897 GGGGAUUGUAACUCACUCCCCCUUUUUGGGGCUUAUCCGAAGCAGUACCUCUACACUCGGGCAAACGAGUGGGGUAGUUCUUACCUAUUGGUACCUGUCGACGUUAAGCUUAUACCCAAA (((((.((.....)).)))))...(((((((((((..((..((((((((....((((((......))))))((((((...))))))...)))).))).)..)).))))).....)))))) ( -42.90) >DroSec_CAF1 24219 119 + 1 GGGGAUUGUAACUCACUCCCCCUU-UUGGGGCUUAUCCGAAGCAGUACCUCUACACUCGGGCAAACGAGUGGGGUAUUUCUUACCUAUUAGUACCUGUCGACGUUAAGCUUAUACCCAAA (((((.((.....)).)))))..(-((((((((((..((..(((((((.....((((((......))))))(((((.....)))))....))).))).)..)).))))).....)))))) ( -40.50) >DroSim_CAF1 24333 119 + 1 GGGGAUUGUAACUCACUCCCCCUU-UUGGGGCUUAUCCGAAGCAGUACCUCUACACUCGGGCAAACGAGUGGGGUAUUUCUUACCUAUUAGUACCUGUCGACGUUAAGCUUAUACCCAAA (((((.((.....)).)))))..(-((((((((((..((..(((((((.....((((((......))))))(((((.....)))))....))).))).)..)).))))).....)))))) ( -40.50) >consensus GGGGAUUGUAACUCACUCCCCCUU_UUGGGGCUUAUCCGAAGCAGUACCUCUACACUCGGGCAAACGAGUGGGGUAUUUCUUACCUAUUAGUACCUGUCGACGUUAAGCUUAUACCCAAA (((((.((.....)).)))))....((((((((((..((..(((((((.....((((((......))))))(((((.....)))))....))).))).)..)).))))).....))))). (-39.77 = -39.77 + 0.00)

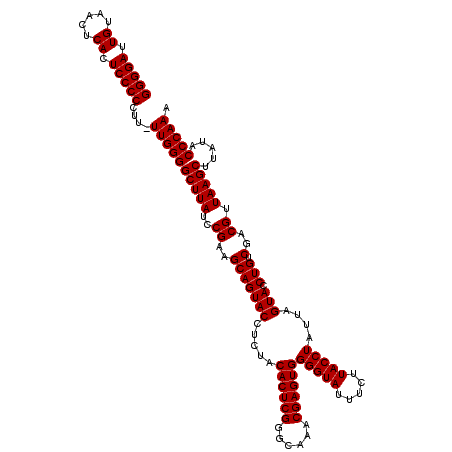
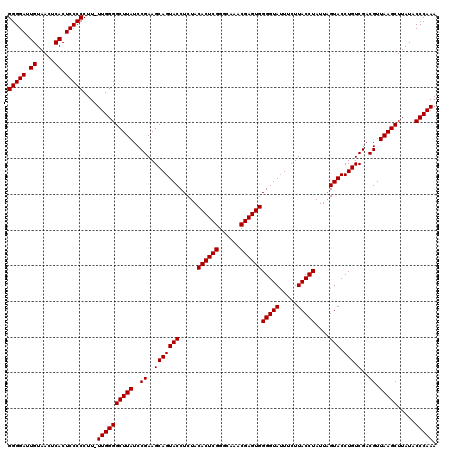
| Location | 21,518,919 – 21,519,039 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -40.33 |
| Consensus MFE | -39.32 |
| Energy contribution | -39.10 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960728 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21518919 120 - 23771897 UUUGGGUAUAAGCUUAACGUCGACAGGUACCAAUAGGUAAGAACUACCCCACUCGUUUGCCCGAGUGUAGAGGUACUGCUUCGGAUAAGCCCCAAAAAGGGGGAGUGAGUUACAAUCCCC ((((((.....(((((...((((..((((((....((((.....)))).((((((......))))))....))))))...)))).)))))))))))...(((((.((.....)).))))) ( -42.20) >DroSec_CAF1 24219 119 - 1 UUUGGGUAUAAGCUUAACGUCGACAGGUACUAAUAGGUAAGAAAUACCCCACUCGUUUGCCCGAGUGUAGAGGUACUGCUUCGGAUAAGCCCCAA-AAGGGGGAGUGAGUUACAAUCCCC ((((((.....(((((...((((..((((((....((((.....)))).((((((......))))))....))))))...)))).))))))))))-)..(((((.((.....)).))))) ( -39.40) >DroSim_CAF1 24333 119 - 1 UUUGGGUAUAAGCUUAACGUCGACAGGUACUAAUAGGUAAGAAAUACCCCACUCGUUUGCCCGAGUGUAGAGGUACUGCUUCGGAUAAGCCCCAA-AAGGGGGAGUGAGUUACAAUCCCC ((((((.....(((((...((((..((((((....((((.....)))).((((((......))))))....))))))...)))).))))))))))-)..(((((.((.....)).))))) ( -39.40) >consensus UUUGGGUAUAAGCUUAACGUCGACAGGUACUAAUAGGUAAGAAAUACCCCACUCGUUUGCCCGAGUGUAGAGGUACUGCUUCGGAUAAGCCCCAA_AAGGGGGAGUGAGUUACAAUCCCC .(((((.....(((((...((((..((((((....((((.....)))).((((((......))))))....))))))...)))).))))))))))....(((((.((.....)).))))) (-39.32 = -39.10 + -0.22)

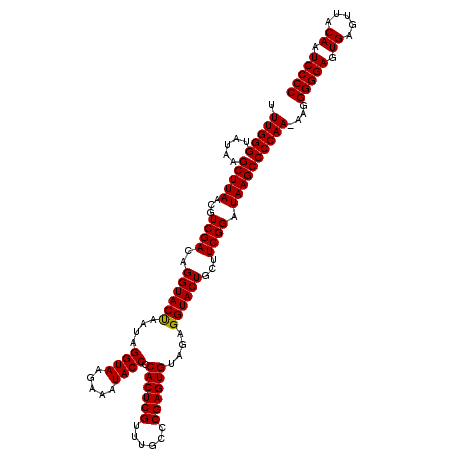
| Location | 21,518,959 – 21,519,076 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -32.03 |
| Consensus MFE | -29.53 |
| Energy contribution | -29.76 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21518959 117 - 23771897 ---AGGCAAAUACCGUUCAUAUCUGAAGGUAUCAAAAUUAUUUGGGUAUAAGCUUAACGUCGACAGGUACCAAUAGGUAAGAACUACCCCACUCGUUUGCCCGAGUGUAGAGGUACUGCU ---.((((..((((((((.((((((..((((((........((((((....))))))........))))))..)))))).))))..(..((((((......))))))..).)))).)))) ( -35.19) >DroSec_CAF1 24258 120 - 1 AUUAGGCAAUUACCGUUCAUAUCUGAAGGUACCACAAUUAUUUGGGUAUAAGCUUAACGUCGACAGGUACUAAUAGGUAAGAAAUACCCCACUCGUUUGCCCGAGUGUAGAGGUACUGCU ....((((..((((.(((.((((((..((((((........((((((....))))))........))))))..)))))).)))...(..((((((......))))))..).)))).)))) ( -32.69) >DroSim_CAF1 24372 119 - 1 AUUAGGCAAUUACCGUUCAUAUAUGAAGGUAC-AAAAUUAUUUGGGUAUAAGCUUAACGUCGACAGGUACUAAUAGGUAAGAAAUACCCCACUCGUUUGCCCGAGUGUAGAGGUACUGCU .((((((...((((.((((....))))))))(-(((....)))).......))))))........((((((....((((.....)))).((((((......))))))....))))))... ( -28.20) >consensus AUUAGGCAAUUACCGUUCAUAUCUGAAGGUACCAAAAUUAUUUGGGUAUAAGCUUAACGUCGACAGGUACUAAUAGGUAAGAAAUACCCCACUCGUUUGCCCGAGUGUAGAGGUACUGCU ....((((..((((.(((.((((((..((((((........((((((....))))))........))))))..)))))).)))...(..((((((......))))))..).)))).)))) (-29.53 = -29.76 + 0.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:08:35 2006