| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
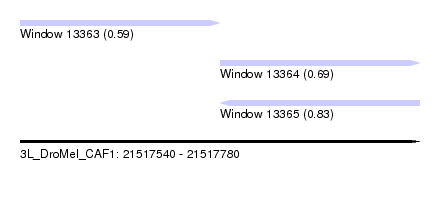
| Location | 21,517,540 – 21,517,780 |
| Length | 240 |
| Max. P | 0.828502 |

| Location | 21,517,540 – 21,517,660 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -25.77 |
| Consensus MFE | -22.50 |
| Energy contribution | -25.50 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21517540 120 + 23771897 UCGACUGCAAUUGCAUUUGAAUGUUUUUCGAGGCUUCGGCCUCUUUUUGUUUAAAUAUAUAAAAGCCAUUAAAUACUAAUCUAUUCUCGAUUGCUCUUCUUCGGCUUCUUCGGGCACCUA ..((..(((((((.....(((((......(((((....)))))(((((((........)))))))................))))).)))))))...))...((..((....))..)).. ( -26.30) >DroSec_CAF1 22778 120 + 1 UCGACUGCAAUUGCAUUUGAAUGUUUUUCGAGGCUUCGGCCUCUUUUUGUUUAAAUAUAUAAAAGCCAUUAAAUACUAAUCUAUUCUCGAUUGCUCUUCUUCGGCUACUUCGGGCACCUA ((((.(((....))).)))).........(((((....))))).((((((........))))))(((((((.....)))).......(((..(((.......)))....))))))..... ( -25.50) >DroSim_CAF1 22891 120 + 1 UCGACUGCAAUUGCAUUUGAAUGUUUUUCGAGGCUUCGGCCUCUUUUUGUUUAAAUAUAUAAAAGCCAUUAAAUACUAAUCUAUUCUCGAUUGCUCUUCUUCGGCUACUUCGGGCACCUA ((((.(((....))).)))).........(((((....))))).((((((........))))))(((((((.....)))).......(((..(((.......)))....))))))..... ( -25.50) >consensus UCGACUGCAAUUGCAUUUGAAUGUUUUUCGAGGCUUCGGCCUCUUUUUGUUUAAAUAUAUAAAAGCCAUUAAAUACUAAUCUAUUCUCGAUUGCUCUUCUUCGGCUACUUCGGGCACCUA ((((.(((....))).)))).........(((((....))))).((((((........))))))(((((((.....)))).......(((..(((.......)))....))))))..... (-22.50 = -25.50 + 3.00)



| Location | 21,517,660 – 21,517,780 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -42.57 |
| Consensus MFE | -40.85 |
| Energy contribution | -40.97 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690112 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21517660 120 + 23771897 AUGACGCGCGUUCAACAUUUAGGCCACCAUCGGGCCAGGGGACCAGUGGGUCCCGUCCAGUGCAAGGAGGCAGUGCCUCCUCCCUCCUAAGUAUGAUUAAUUAAUAAUGCAGGUAAGCAU ..(((................((((.......))))..((((((....)))))))))..((((.(((((((...)))))))....(((..((((.(((....))).)))))))...)))) ( -39.60) >DroSec_CAF1 22898 120 + 1 AUGACGCGCGGCCAACAUUUAGGCCACCAUCGGGCCAGGGGACCAGUGGGUCCCGUCCAGUGCAAGGAGGCAGUGUCUCCUCCCUCCUAAGUAUGAUUAAUUAAUAAUGCAGGUAAGCAU ...((((((((((........))))......((((...((((((....)))))))))).)))).((((((.((......)).))))))..))..............((((......)))) ( -43.80) >DroSim_CAF1 23011 120 + 1 AUGACGCGCGGCCAACAUUUAGGCCACCAUCGGGCCAGGGGACCAGUGGGUCCCGUCCAGUGCAAGGAGGCAGUGCCUCCUCCCUCCUAAGUAUGAUUAAUUAAUAAUGCAGGUAAGCAU .....((((((((........))))......((((...((((((....)))))))))).)))).(((((((...)))))))....(((..((((.(((....))).)))))))....... ( -44.30) >consensus AUGACGCGCGGCCAACAUUUAGGCCACCAUCGGGCCAGGGGACCAGUGGGUCCCGUCCAGUGCAAGGAGGCAGUGCCUCCUCCCUCCUAAGUAUGAUUAAUUAAUAAUGCAGGUAAGCAU ...((((((((((........))))......((((...((((((....)))))))))).)))).((((((.((......)).))))))..))..............((((......)))) (-40.85 = -40.97 + 0.11)


| Location | 21,517,660 – 21,517,780 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -44.77 |
| Consensus MFE | -42.33 |
| Energy contribution | -43.33 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.828502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21517660 120 - 23771897 AUGCUUACCUGCAUUAUUAAUUAAUCAUACUUAGGAGGGAGGAGGCACUGCCUCCUUGCACUGGACGGGACCCACUGGUCCCCUGGCCCGAUGGUGGCCUAAAUGUUGAACGCGCGUCAU ((((......)))).................(((..(.((((((((...)))))))).).)))(((((((((....))))))..(((((....).))))................))).. ( -40.00) >DroSec_CAF1 22898 120 - 1 AUGCUUACCUGCAUUAUUAAUUAAUCAUACUUAGGAGGGAGGAGACACUGCCUCCUUGCACUGGACGGGACCCACUGGUCCCCUGGCCCGAUGGUGGCCUAAAUGUUGGCCGCGCGUCAU ..(((..(((((((((.....)))........((((((.((......)).))))))))))..))..((((((....))))))..)))..((((((((((........)))))).)))).. ( -45.40) >DroSim_CAF1 23011 120 - 1 AUGCUUACCUGCAUUAUUAAUUAAUCAUACUUAGGAGGGAGGAGGCACUGCCUCCUUGCACUGGACGGGACCCACUGGUCCCCUGGCCCGAUGGUGGCCUAAAUGUUGGCCGCGCGUCAU ((((......))))...................((.(.((((((((...)))))))).).(..(..((((((....)))))))..).))((((((((((........)))))).)))).. ( -48.90) >consensus AUGCUUACCUGCAUUAUUAAUUAAUCAUACUUAGGAGGGAGGAGGCACUGCCUCCUUGCACUGGACGGGACCCACUGGUCCCCUGGCCCGAUGGUGGCCUAAAUGUUGGCCGCGCGUCAU ((((......))))...................((.(.((((((((...)))))))).).(..(..((((((....)))))))..).))((((((((((........)))))).)))).. (-42.33 = -43.33 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:08:26 2006