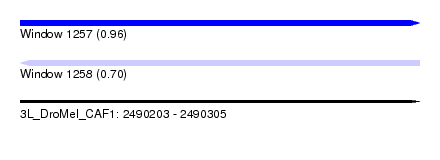
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,490,203 – 2,490,305 |
| Length | 102 |
| Max. P | 0.958438 |

| Location | 2,490,203 – 2,490,305 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 85.03 |
| Mean single sequence MFE | -47.52 |
| Consensus MFE | -40.75 |
| Energy contribution | -41.67 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
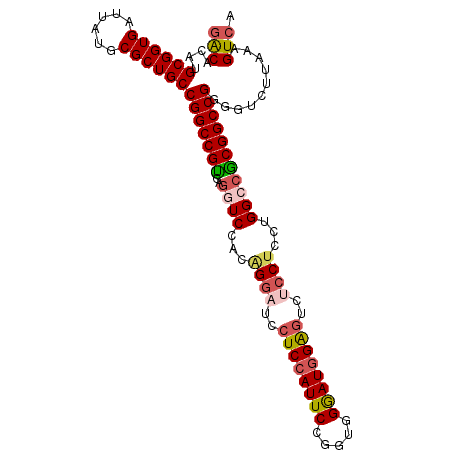
>3L_DroMel_CAF1 2490203 102 + 23771897 GACACAUGCGGUGAUUAUGCGCUGCCGGCCGUCAGGUCCACAGGAUCCUCCAUUCCGGUGGGAUGGAGUCUCCUUCUGGCCGCGGCCGGGGUCUUAAAGUCA (((....((((((......))))))(((((((..((.(((.((((..(((((((((...)))))))))..))))..))))))))))))..........))). ( -50.20) >DroSec_CAF1 113614 102 + 1 GACACAUGCGGUGAUUAUGCGCUGCCGGCCGUCAGGUCCACAGGAUCCUCCAUUCCGGUGGGAUGGAGUCUCCUCCUGGCCGCGGCCGGGGUCUUAAAGUCA (((....((((((......))))))(((((((..((((...((((..(((((((((...)))))))))..))))...)))))))))))..........))). ( -49.90) >DroSim_CAF1 108589 102 + 1 GACACAUGCGGUGAUUAUGCGCUGCCGGCCGUCAGGUCCACAGGAUCCUCCAUUCCAGUGGGAUGGAGUCUCCUCCUGGCCGCGGCCGGGGUCUUAAAGUCA (((....((((((......))))))(((((((..((((...((((..(((((((((...)))))))))..))))...)))))))))))..........))). ( -49.90) >DroEre_CAF1 113852 102 + 1 GACACAUGCGGUGAUUAUGCGCUGCCGGCCGCCGGGUCCACGGGAUCCUCCAUUCGGUUGGGAUGGAGUCUCCUUGUGGCCGCGGCCGGGGUCUUAAAGUCA (((....((((((......))))))(((((((..((.((((((((..((((((((.....))))))))..))).))))))))))))))..........))). ( -53.20) >DroYak_CAF1 111945 102 + 1 GACACAUGCGGUGAUUAUGCGCUGCCGGCCGUCGGAUCCACGGACUCCUCCAUUCCGGUGGAAUGGAUUAUCCUUGUGGCCACGGCCGGGGUCUUAAAGUCA (((....((((((......))))))(((((((.((..(((((((....((((((((...))))))))...)))..)))))))))))))..........))). ( -44.70) >DroAna_CAF1 94083 89 + 1 UGCACAUGCGGUGAAUAUGCGCUGCCGGCCGUCGACUCCCGGGGCGCCUCU-------------UGGGCACCCACCUGGCCACGGCCGGGGUCUUAAAGUCA .((....((((((......))))))..))....((((....(((.(((...-------------..))).))).((((((....)))))).......)))). ( -37.20) >consensus GACACAUGCGGUGAUUAUGCGCUGCCGGCCGUCAGGUCCACAGGAUCCUCCAUUCCGGUGGGAUGGAGUCUCCUCCUGGCCGCGGCCGGGGUCUUAAAGUCA (((....((((((......))))))(((((((..((((...((((..((((((((.....))))))))..))))...)))))))))))..........))). (-40.75 = -41.67 + 0.92)

| Location | 2,490,203 – 2,490,305 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 85.03 |
| Mean single sequence MFE | -42.45 |
| Consensus MFE | -33.26 |
| Energy contribution | -33.67 |
| Covariance contribution | 0.41 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2490203 102 - 23771897 UGACUUUAAGACCCCGGCCGCGGCCAGAAGGAGACUCCAUCCCACCGGAAUGGAGGAUCCUGUGGACCUGACGGCCGGCAGCGCAUAAUCACCGCAUGUGUC .............(((((((((((((..((((..((((((((....)).))))))..)))).))).)).).)))))))..((((((.........)))))). ( -43.50) >DroSec_CAF1 113614 102 - 1 UGACUUUAAGACCCCGGCCGCGGCCAGGAGGAGACUCCAUCCCACCGGAAUGGAGGAUCCUGUGGACCUGACGGCCGGCAGCGCAUAAUCACCGCAUGUGUC .............(((((((((((((.(.(((..((((((((....)).))))))..)))).))).)).).)))))))..((((((.........)))))). ( -42.40) >DroSim_CAF1 108589 102 - 1 UGACUUUAAGACCCCGGCCGCGGCCAGGAGGAGACUCCAUCCCACUGGAAUGGAGGAUCCUGUGGACCUGACGGCCGGCAGCGCAUAAUCACCGCAUGUGUC .............(((((((((((((.(.(((..((((((((....)).))))))..)))).))).)).).)))))))..((((((.........)))))). ( -42.30) >DroEre_CAF1 113852 102 - 1 UGACUUUAAGACCCCGGCCGCGGCCACAAGGAGACUCCAUCCCAACCGAAUGGAGGAUCCCGUGGACCCGGCGGCCGGCAGCGCAUAAUCACCGCAUGUGUC .............((((((((((((((..(((..((((((.(.....).))))))..))).)))).))..))))))))..((((((.........)))))). ( -43.00) >DroYak_CAF1 111945 102 - 1 UGACUUUAAGACCCCGGCCGUGGCCACAAGGAUAAUCCAUUCCACCGGAAUGGAGGAGUCCGUGGAUCCGACGGCCGGCAGCGCAUAAUCACCGCAUGUGUC .............((((((((((((((..((((..((((((((...))))))))...))))))))..)).))))))))..((((((.........)))))). ( -42.30) >DroAna_CAF1 94083 89 - 1 UGACUUUAAGACCCCGGCCGUGGCCAGGUGGGUGCCCA-------------AGAGGCGCCCCGGGAGUCGACGGCCGGCAGCGCAUAUUCACCGCAUGUGCA .............((((((((((((..(.(((((((..-------------...))))))))..).))).))))))))..((((((.........)))))). ( -41.20) >consensus UGACUUUAAGACCCCGGCCGCGGCCAGAAGGAGACUCCAUCCCACCGGAAUGGAGGAUCCCGUGGACCCGACGGCCGGCAGCGCAUAAUCACCGCAUGUGUC .............(((((((((((((.(.(((..((((((.(.....).))))))..)))).)))..))).)))))))..((((((.........)))))). (-33.26 = -33.67 + 0.41)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:56 2006