| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,514,868 – 21,515,053 |
| Length | 185 |
| Max. P | 0.988536 |

| Location | 21,514,868 – 21,514,973 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 91.23 |
| Mean single sequence MFE | -46.10 |
| Consensus MFE | -44.27 |
| Energy contribution | -44.17 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21514868 105 - 23771897 UGUUACUCCUCGUGAGAGGAGAGGGCCAGGGUGAUGGCCCAGGGCGAUCAUGUACGACCUGGAGAGACUGGAGUUGCGGCCGCCUCGAUGGCUC---------AAAUGGCAAAG (((((((((((....)))))).((((((((((....)))).(((((..(.....(((((..(.....)..).)))).)..)))))...))))))---------...)))))... ( -44.40) >DroSec_CAF1 19988 114 - 1 UGUUGCUCCUCUGGAGAGGAGAGGGCCAGGGCGAUGGCCCGGGGCGAUCAUGUACGACCUGGAGAGACUGGAGUUGUGGCCGCCUCGAUGGCUCAAAUGGCUCAAAUGGCAAAG .....((((((....)))))).((((((((((....))))((((((..(....((((((..(.....)..).))))))..))))))..)))))).....(((.....))).... ( -48.00) >DroSim_CAF1 20098 114 - 1 UGUUGCUCCUCUGGAGAGGAGAGGGCCAGGGCGAUGGCCCGGGGCGAUCAUGUACGACCUGGAGAGACUGGAGUUGCGGCCGCCUCGAUGGCUCAAAUGGCUCAAAUGGCAAAG .....((((((....)))))).((((((((((....))))((((((..(.....(((((..(.....)..).)))).)..))))))..)))))).....(((.....))).... ( -45.90) >consensus UGUUGCUCCUCUGGAGAGGAGAGGGCCAGGGCGAUGGCCCGGGGCGAUCAUGUACGACCUGGAGAGACUGGAGUUGCGGCCGCCUCGAUGGCUCAAAUGGCUCAAAUGGCAAAG (((((((((((....)))))).((((((((((....))))((((((..(.....(((((..(.....)..).)))).)..))))))..))))))............)))))... (-44.27 = -44.17 + -0.11)



| Location | 21,514,893 – 21,515,013 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.66 |
| Mean single sequence MFE | -52.60 |
| Consensus MFE | -50.43 |
| Energy contribution | -50.77 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21514893 120 + 23771897 GCCGCAACUCCAGUCUCUCCAGGUCGUACAUGAUCGCCCUGGGCCAUCACCCUGGCCCUCUCCUCUCACGAGGAGUAACAGAAGAUGGGGUUGGAGAUGGGCAGGAGGUGGGCCGCGGCA (((((...(((.((((((((((((((....)))))(((((((((((......)))))).((((((....))))))...........)))))))))))..))).)))((....))))))). ( -57.80) >DroSec_CAF1 20022 108 + 1 GCCACAACUCCAGUCUCUCCAGGUCGUACAUGAUCGCCCCGGGCCAUCGCCCUGGCCCUCUCCUCUCCAGAGGAGCAACAGAAGA------------UGGGCAGGAGGUGGGCCGCGGCA (((.(...((((...(((((.(((((....)))))((((.((((((......)))))).((((((....))))))..........------------.)))).)))))))))..).))). ( -47.30) >DroSim_CAF1 20132 108 + 1 GCCGCAACUCCAGUCUCUCCAGGUCGUACAUGAUCGCCCCGGGCCAUCGCCCUGGCCCUCUCCUCUCCAGAGGAGCAACAGAAGA------------UGGGCAGGAGGUGGGCCGCGGCA (((((...((((...(((((.(((((....)))))((((.((((((......)))))).((((((....))))))..........------------.)))).)))))))))..))))). ( -52.70) >consensus GCCGCAACUCCAGUCUCUCCAGGUCGUACAUGAUCGCCCCGGGCCAUCGCCCUGGCCCUCUCCUCUCCAGAGGAGCAACAGAAGA____________UGGGCAGGAGGUGGGCCGCGGCA (((((...((((...(((((.(((((....)))))((((.((((((......)))))).((((((....)))))).......................)))).)))))))))..))))). (-50.43 = -50.77 + 0.33)

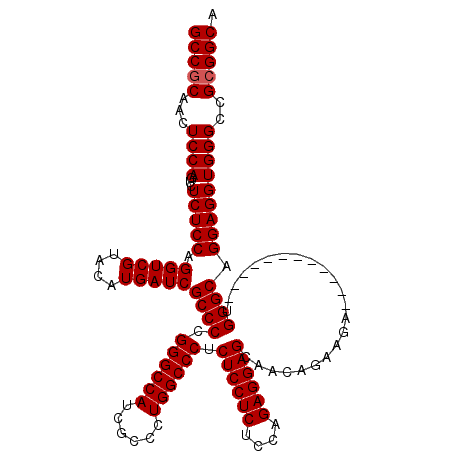

| Location | 21,514,893 – 21,515,013 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.66 |
| Mean single sequence MFE | -53.50 |
| Consensus MFE | -53.72 |
| Energy contribution | -53.50 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.98 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21514893 120 - 23771897 UGCCGCGGCCCACCUCCUGCCCAUCUCCAACCCCAUCUUCUGUUACUCCUCGUGAGAGGAGAGGGCCAGGGUGAUGGCCCAGGGCGAUCAUGUACGACCUGGAGAGACUGGAGUUGCGGC .((((((((((((((((.((((.......................((((((....)))))).((((((......)))))).))))(.((......)).).)))).)..))).)))))))) ( -56.80) >DroSec_CAF1 20022 108 - 1 UGCCGCGGCCCACCUCCUGCCCA------------UCUUCUGUUGCUCCUCUGGAGAGGAGAGGGCCAGGGCGAUGGCCCGGGGCGAUCAUGUACGACCUGGAGAGACUGGAGUUGUGGC .((((((((((((((((.((((.------------..........((((((....)))))).((((((......)))))).))))(.((......)).).)))).)..))).)))))))) ( -50.90) >DroSim_CAF1 20132 108 - 1 UGCCGCGGCCCACCUCCUGCCCA------------UCUUCUGUUGCUCCUCUGGAGAGGAGAGGGCCAGGGCGAUGGCCCGGGGCGAUCAUGUACGACCUGGAGAGACUGGAGUUGCGGC .((((((((((((((((.((((.------------..........((((((....)))))).((((((......)))))).))))(.((......)).).)))).)..))).)))))))) ( -52.80) >consensus UGCCGCGGCCCACCUCCUGCCCA____________UCUUCUGUUGCUCCUCUGGAGAGGAGAGGGCCAGGGCGAUGGCCCGGGGCGAUCAUGUACGACCUGGAGAGACUGGAGUUGCGGC .((((((((((((((((.((((.......................((((((....)))))).((((((......)))))).))))(.((......)).).)))).)..))).)))))))) (-53.72 = -53.50 + -0.22)


| Location | 21,514,933 – 21,515,053 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.23 |
| Mean single sequence MFE | -53.06 |
| Consensus MFE | -51.09 |
| Energy contribution | -50.87 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.936750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21514933 120 + 23771897 GGGCCAUCACCCUGGCCCUCUCCUCUCACGAGGAGUAACAGAAGAUGGGGUUGGAGAUGGGCAGGAGGUGGGCCGCGGCACAGGCGUUGCGUCUGGCACGGGCUAGUGGCCAUAAUUCUC .((((((((((((.(((((((((.(((((.....))..((.....)))))..))))).)))).)).))))((((.((((.((((((...)))))))).)))))).))))))......... ( -55.30) >DroSec_CAF1 20062 108 + 1 GGGCCAUCGCCCUGGCCCUCUCCUCUCCAGAGGAGCAACAGAAGA------------UGGGCAGGAGGUGGGCCGCGGCACAGGCGUUGGGUCUGGCACGGGCUAGUGGCCAUAAUUCUC .((((((.((((.(((((((((((.((((................------------)))).)))))).)))))...((.(((((.....)))))))..))))..))))))......... ( -51.59) >DroSim_CAF1 20172 108 + 1 GGGCCAUCGCCCUGGCCCUCUCCUCUCCAGAGGAGCAACAGAAGA------------UGGGCAGGAGGUGGGCCGCGGCACAGGCGUUGCGUCUGGCACGGGCUAGUGGCCAUAAUUCUC .((((((.((((.(((((((((((.((((................------------)))).)))))).)))))...((.((((((...))))))))..))))..))))))......... ( -52.29) >consensus GGGCCAUCGCCCUGGCCCUCUCCUCUCCAGAGGAGCAACAGAAGA____________UGGGCAGGAGGUGGGCCGCGGCACAGGCGUUGCGUCUGGCACGGGCUAGUGGCCAUAAUUCUC .((((((((((((.((((.((((((....))))))...(....)..............)))).)).))))((((.((((.(((((.....))))))).)))))).))))))......... (-51.09 = -50.87 + -0.22)

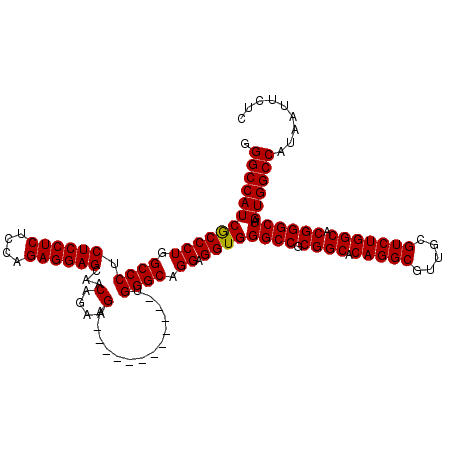
| Location | 21,514,933 – 21,515,053 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.23 |
| Mean single sequence MFE | -45.23 |
| Consensus MFE | -43.49 |
| Energy contribution | -43.27 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21514933 120 - 23771897 GAGAAUUAUGGCCACUAGCCCGUGCCAGACGCAACGCCUGUGCCGCGGCCCACCUCCUGCCCAUCUCCAACCCCAUCUUCUGUUACUCCUCGUGAGAGGAGAGGGCCAGGGUGAUGGCCC .........(((((...(((((.(((((.((...)).))).)))).))).(((((...((((......(((..........))).((((((....)))))).))))..))))).))))). ( -47.90) >DroSec_CAF1 20062 108 - 1 GAGAAUUAUGGCCACUAGCCCGUGCCAGACCCAACGCCUGUGCCGCGGCCCACCUCCUGCCCA------------UCUUCUGUUGCUCCUCUGGAGAGGAGAGGGCCAGGGCGAUGGCCC .........(((((...(((((((((((.........))).).)))(((((..(((((...((------------........))((((...))))))))).))))).))))..))))). ( -43.30) >DroSim_CAF1 20172 108 - 1 GAGAAUUAUGGCCACUAGCCCGUGCCAGACGCAACGCCUGUGCCGCGGCCCACCUCCUGCCCA------------UCUUCUGUUGCUCCUCUGGAGAGGAGAGGGCCAGGGCGAUGGCCC .........(((((...(((((((((((.((...)).))).).)))(((((..(((((...((------------........))((((...))))))))).))))).))))..))))). ( -44.50) >consensus GAGAAUUAUGGCCACUAGCCCGUGCCAGACGCAACGCCUGUGCCGCGGCCCACCUCCUGCCCA____________UCUUCUGUUGCUCCUCUGGAGAGGAGAGGGCCAGGGCGAUGGCCC .........(((((...(((((((((((.(.....).))).).)))(((((..(....)..........................((((((....)))))).))))).))))..))))). (-43.49 = -43.27 + -0.22)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:08:19 2006