
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,511,418 – 21,511,738 |
| Length | 320 |
| Max. P | 0.992552 |

| Location | 21,511,418 – 21,511,538 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -51.57 |
| Consensus MFE | -50.45 |
| Energy contribution | -50.23 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.02 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21511418 120 + 23771897 ACGGACCACCCCGAAAUUGACUGGAGUGCGAUGCGUGCCGAGUGGUUGCAGUUCGAAUUUGUAAUUACAAUUCGUGGCGCCUUGCCGGGGAAGUUCUGCUGGGGGAGCCCCCAGCACUGG ..((((..(((((......((....))((((.((((..(((((((((((((.......))))))))))...)))..)))).)))))))))..))))((((((((....)))))))).... ( -52.00) >DroSec_CAF1 16488 120 + 1 ACGGACCACCCCGAAAUUGACUGGAGUGCGAUGCGUGCCAAGUGGUUGCAGUUCGAAUUUGUAAUUACAAUUCGUGGCGCCUUGCCGGGGAAGUUCUGCUGGGGGAACCCCCAGCACUGG ..((((..(((((......((....))((((.(.((((((.((((((((((.......))))))))))......))))))))))))))))..))))((((((((....)))))))).... ( -50.70) >DroSim_CAF1 16115 120 + 1 ACGGACCACCCCGAAAUUGACUGGAGUGCGAUGCGUGCCGAGUGGUUGCAGUUCGAAUUUGUAAUUACAAUUCGUGGCGCCUUGCCGGGGAAGUUCUGCUGGGGGAACCCCCAGCACUGG ..((((..(((((......((....))((((.((((..(((((((((((((.......))))))))))...)))..)))).)))))))))..))))((((((((....)))))))).... ( -52.00) >consensus ACGGACCACCCCGAAAUUGACUGGAGUGCGAUGCGUGCCGAGUGGUUGCAGUUCGAAUUUGUAAUUACAAUUCGUGGCGCCUUGCCGGGGAAGUUCUGCUGGGGGAACCCCCAGCACUGG ..((((..(((((......((....))((((.(.((((((.((((((((((.......))))))))))......))))))))))))))))..))))((((((((....)))))))).... (-50.45 = -50.23 + -0.22)

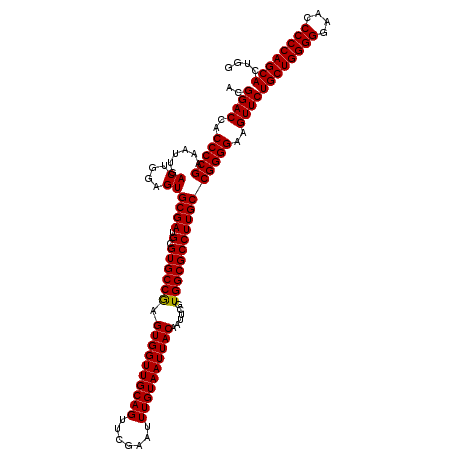
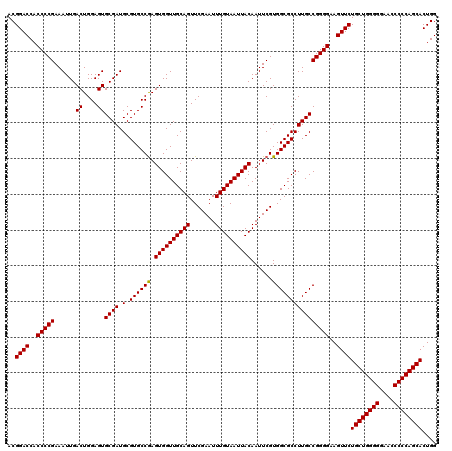
| Location | 21,511,418 – 21,511,538 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -41.00 |
| Consensus MFE | -40.80 |
| Energy contribution | -40.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.622964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21511418 120 - 23771897 CCAGUGCUGGGGGCUCCCCCAGCAGAACUUCCCCGGCAAGGCGCCACGAAUUGUAAUUACAAAUUCGAACUGCAACCACUCGGCACGCAUCGCACUCCAGUCAAUUUCGGGGUGGUCCGU ....((((((((....))))))))(.(((.((((((...((((((.((((((((....)).))))))...((....))...)))..((...))......)))....)))))).))).).. ( -40.90) >DroSec_CAF1 16488 120 - 1 CCAGUGCUGGGGGUUCCCCCAGCAGAACUUCCCCGGCAAGGCGCCACGAAUUGUAAUUACAAAUUCGAACUGCAACCACUUGGCACGCAUCGCACUCCAGUCAAUUUCGGGGUGGUCCGU ....((((((((....))))))))(.(((.((((((...(((((((((((((((....)).))))))...((....))..))))..((...))......)))....)))))).))).).. ( -41.20) >DroSim_CAF1 16115 120 - 1 CCAGUGCUGGGGGUUCCCCCAGCAGAACUUCCCCGGCAAGGCGCCACGAAUUGUAAUUACAAAUUCGAACUGCAACCACUCGGCACGCAUCGCACUCCAGUCAAUUUCGGGGUGGUCCGU ....((((((((....))))))))(.(((.((((((...((((((.((((((((....)).))))))...((....))...)))..((...))......)))....)))))).))).).. ( -40.90) >consensus CCAGUGCUGGGGGUUCCCCCAGCAGAACUUCCCCGGCAAGGCGCCACGAAUUGUAAUUACAAAUUCGAACUGCAACCACUCGGCACGCAUCGCACUCCAGUCAAUUUCGGGGUGGUCCGU ....((((((((....))))))))(.(((.((((((...((((((.((((((((....)).))))))...((....))...)))..((...))......)))....)))))).))).).. (-40.80 = -40.80 + 0.00)



| Location | 21,511,458 – 21,511,578 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -54.23 |
| Consensus MFE | -54.45 |
| Energy contribution | -54.23 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -4.48 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.984925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21511458 120 + 23771897 AGUGGUUGCAGUUCGAAUUUGUAAUUACAAUUCGUGGCGCCUUGCCGGGGAAGUUCUGCUGGGGGAGCCCCCAGCACUGGCUGAAAACAAUUUUCUCCAGGUUUUCUCCGUGGUAAGGGC .((((((((((.......))))))))))........((.((((((((.(((.....((((((((....))))))))((((..((((.....)))).))))......))).)))))))))) ( -52.90) >DroSec_CAF1 16528 120 + 1 AGUGGUUGCAGUUCGAAUUUGUAAUUACAAUUCGUGGCGCCUUGCCGGGGAAGUUCUGCUGGGGGAACCCCCAGCACUGGCUGAAAACAAUUUUCUCCAGGUUUUCUCCGUGGCAAGGGC .((((((((((.......))))))))))........((.((((((((.(((.....((((((((....))))))))((((..((((.....)))).))))......))).)))))))))) ( -54.90) >DroSim_CAF1 16155 120 + 1 AGUGGUUGCAGUUCGAAUUUGUAAUUACAAUUCGUGGCGCCUUGCCGGGGAAGUUCUGCUGGGGGAACCCCCAGCACUGGCUGAAAACAAUUUUCUCCAGGUUUUCUCCGUGGCAAGGGC .((((((((((.......))))))))))........((.((((((((.(((.....((((((((....))))))))((((..((((.....)))).))))......))).)))))))))) ( -54.90) >consensus AGUGGUUGCAGUUCGAAUUUGUAAUUACAAUUCGUGGCGCCUUGCCGGGGAAGUUCUGCUGGGGGAACCCCCAGCACUGGCUGAAAACAAUUUUCUCCAGGUUUUCUCCGUGGCAAGGGC .((((((((((.......))))))))))........((.((((((((.(((.....((((((((....))))))))((((..((((.....)))).))))......))).)))))))))) (-54.45 = -54.23 + -0.22)

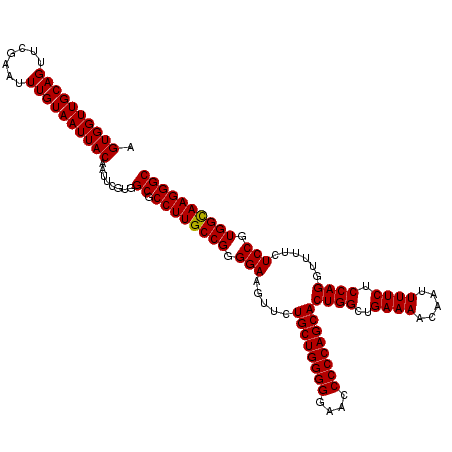

| Location | 21,511,458 – 21,511,578 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -44.20 |
| Consensus MFE | -44.23 |
| Energy contribution | -44.57 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.65 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21511458 120 - 23771897 GCCCUUACCACGGAGAAAACCUGGAGAAAAUUGUUUUCAGCCAGUGCUGGGGGCUCCCCCAGCAGAACUUCCCCGGCAAGGCGCCACGAAUUGUAAUUACAAAUUCGAACUGCAACCACU ((((((.((..((((.....((((.((((.....))))..))))((((((((....))))))))....))))..)).)))).))..((((((((....)).))))))............. ( -39.80) >DroSec_CAF1 16528 120 - 1 GCCCUUGCCACGGAGAAAACCUGGAGAAAAUUGUUUUCAGCCAGUGCUGGGGGUUCCCCCAGCAGAACUUCCCCGGCAAGGCGCCACGAAUUGUAAUUACAAAUUCGAACUGCAACCACU (((((((((..((((.....((((.((((.....))))..))))((((((((....))))))))....))))..))))))).))..((((((((....)).))))))............. ( -46.40) >DroSim_CAF1 16155 120 - 1 GCCCUUGCCACGGAGAAAACCUGGAGAAAAUUGUUUUCAGCCAGUGCUGGGGGUUCCCCCAGCAGAACUUCCCCGGCAAGGCGCCACGAAUUGUAAUUACAAAUUCGAACUGCAACCACU (((((((((..((((.....((((.((((.....))))..))))((((((((....))))))))....))))..))))))).))..((((((((....)).))))))............. ( -46.40) >consensus GCCCUUGCCACGGAGAAAACCUGGAGAAAAUUGUUUUCAGCCAGUGCUGGGGGUUCCCCCAGCAGAACUUCCCCGGCAAGGCGCCACGAAUUGUAAUUACAAAUUCGAACUGCAACCACU (((((((((..((((.....((((.((((.....))))..))))((((((((....))))))))....))))..))))))).))..((((((((....)).))))))............. (-44.23 = -44.57 + 0.33)


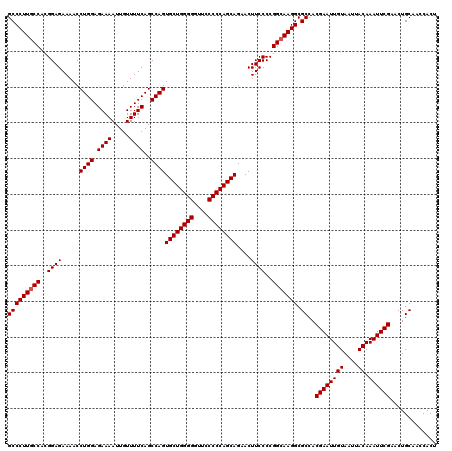
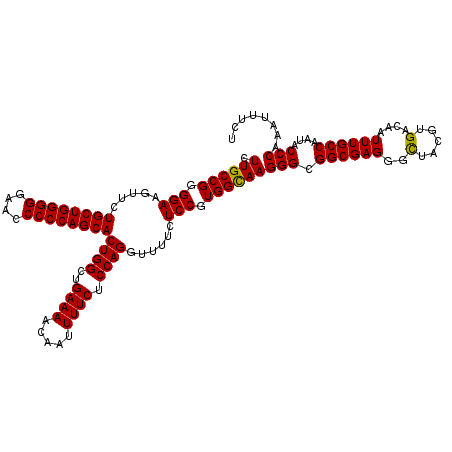
| Location | 21,511,498 – 21,511,618 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -47.47 |
| Consensus MFE | -46.94 |
| Energy contribution | -46.50 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21511498 120 + 23771897 CUUGCCGGGGAAGUUCUGCUGGGGGAGCCCCCAGCACUGGCUGAAAACAAUUUUCUCCAGGUUUUCUCCGUGGUAAGGGCGGCGAGGGCUACGUGACAAUUUGCCAAUACCCAAAUUUCU .((((((.(((.....((((((((....))))))))((((..((((.....)))).))))......))).))))))(((.((((((..(.....)....))))))....)))........ ( -45.80) >DroSec_CAF1 16568 120 + 1 CUUGCCGGGGAAGUUCUGCUGGGGGAACCCCCAGCACUGGCUGAAAACAAUUUUCUCCAGGUUUUCUCCGUGGCAAGGGCGGCGAGGGUUACGUGACAAUUUGCCAAUACCCAAAUUUCU .((((((.(((.....((((((((....))))))))((((..((((.....)))).))))......))).))))))(((.((((((.(((....)))..))))))....)))........ ( -48.80) >DroSim_CAF1 16195 120 + 1 CUUGCCGGGGAAGUUCUGCUGGGGGAACCCCCAGCACUGGCUGAAAACAAUUUUCUCCAGGUUUUCUCCGUGGCAAGGGCGGCGAGGGCUACGUGACAAUUUGCCAAUACCCAAAUUUCU .((((((.(((.....((((((((....))))))))((((..((((.....)))).))))......))).))))))(((.((((((..(.....)....))))))....)))........ ( -47.80) >consensus CUUGCCGGGGAAGUUCUGCUGGGGGAACCCCCAGCACUGGCUGAAAACAAUUUUCUCCAGGUUUUCUCCGUGGCAAGGGCGGCGAGGGCUACGUGACAAUUUGCCAAUACCCAAAUUUCU .((((((.(((.....((((((((....))))))))((((..((((.....)))).))))......))).))))))(((.((((((..(.....)....))))))....)))........ (-46.94 = -46.50 + -0.44)


| Location | 21,511,498 – 21,511,618 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -41.70 |
| Consensus MFE | -41.15 |
| Energy contribution | -41.27 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21511498 120 - 23771897 AGAAAUUUGGGUAUUGGCAAAUUGUCACGUAGCCCUCGCCGCCCUUACCACGGAGAAAACCUGGAGAAAAUUGUUUUCAGCCAGUGCUGGGGGCUCCCCCAGCAGAACUUCCCCGGCAAG ........((((..((((.....))))....))))..((((..........((.(((...((((.((((.....))))..))))((((((((....))))))))....)))))))))... ( -38.90) >DroSec_CAF1 16568 120 - 1 AGAAAUUUGGGUAUUGGCAAAUUGUCACGUAACCCUCGCCGCCCUUGCCACGGAGAAAACCUGGAGAAAAUUGUUUUCAGCCAGUGCUGGGGGUUCCCCCAGCAGAACUUCCCCGGCAAG ........((((((((((.....)))).)).))))........((((((..((((.....((((.((((.....))))..))))((((((((....))))))))....))))..)))))) ( -43.00) >DroSim_CAF1 16195 120 - 1 AGAAAUUUGGGUAUUGGCAAAUUGUCACGUAGCCCUCGCCGCCCUUGCCACGGAGAAAACCUGGAGAAAAUUGUUUUCAGCCAGUGCUGGGGGUUCCCCCAGCAGAACUUCCCCGGCAAG ........((((..((((.....))))....))))........((((((..((((.....((((.((((.....))))..))))((((((((....))))))))....))))..)))))) ( -43.20) >consensus AGAAAUUUGGGUAUUGGCAAAUUGUCACGUAGCCCUCGCCGCCCUUGCCACGGAGAAAACCUGGAGAAAAUUGUUUUCAGCCAGUGCUGGGGGUUCCCCCAGCAGAACUUCCCCGGCAAG ........((((..((((.....))))....))))........((((((..((((.....((((.((((.....))))..))))((((((((....))))))))....))))..)))))) (-41.15 = -41.27 + 0.11)

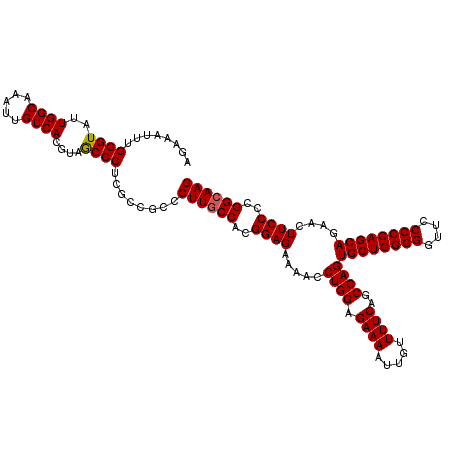

| Location | 21,511,578 – 21,511,698 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -44.23 |
| Consensus MFE | -43.11 |
| Energy contribution | -43.00 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21511578 120 + 23771897 GGCGAGGGCUACGUGACAAUUUGCCAAUACCCAAAUUUCUCCGUGGACCAGGAACGCCUUCUCCGAUGGCGUAAAAAACGAAUAGCCCGUGGGCUUGUUUUUUACCCACGAGGCGAGCCA (((..((.(((((.((.((((((........))))))..))))))).)).....(((((....((.(((.((((((((((...((((....)))))))))))))))))))))))).))). ( -45.60) >DroSec_CAF1 16648 120 + 1 GGCGAGGGUUACGUGACAAUUUGCCAAUACCCAAAUUUCUCCGUGGACCAGGAACGCCUCCUCCGAUGGCGUAAAAAACGAAUAGCCCGUGGGCUUGUUUUUUACCCACGAGGCGAGCCA (((...(((((((.((.((((((........))))))..))))).)))).....((((((....(.(((.((((((((((...((((....)))))))))))))))))))))))).))). ( -43.90) >DroSim_CAF1 16275 120 + 1 GGCGAGGGCUACGUGACAAUUUGCCAAUACCCAAAUUUCUCCGUGGACCAGGAACGCCUCCUCCGAUGGCGUAAAAAACGAAUAGCCCGUGGGCUUGUUUUUUACCGACGAGGCGAGCCA (((..((.(((((.((.((((((........))))))..))))))).)).....((((((.((.....(.((((((((((...((((....))))))))))))))))).)))))).))). ( -43.20) >consensus GGCGAGGGCUACGUGACAAUUUGCCAAUACCCAAAUUUCUCCGUGGACCAGGAACGCCUCCUCCGAUGGCGUAAAAAACGAAUAGCCCGUGGGCUUGUUUUUUACCCACGAGGCGAGCCA (((..((.(((((.((.((((((........))))))..))))))).)).....((((((....(.(((.((((((((((...((((....)))))))))))))))))))))))).))). (-43.11 = -43.00 + -0.11)



| Location | 21,511,618 – 21,511,738 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -39.87 |
| Consensus MFE | -38.55 |
| Energy contribution | -38.67 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21511618 120 + 23771897 CCGUGGACCAGGAACGCCUUCUCCGAUGGCGUAAAAAACGAAUAGCCCGUGGGCUUGUUUUUUACCCACGAGGCGAGCCAACAUGUGGUAACCCAUAAUUUACGCCUAGGGAUGUUAUGA ((.(((....((..(((((....((.(((.((((((((((...((((....))))))))))))))))))))))))..))....(((((....)))))........))).))......... ( -40.50) >DroSec_CAF1 16688 120 + 1 CCGUGGACCAGGAACGCCUCCUCCGAUGGCGUAAAAAACGAAUAGCCCGUGGGCUUGUUUUUUACCCACGAGGCGAGCCAACAUGUGGUAACCCAUAAUUUACGCCUAGGGAUGUUAUGA ((.(((....((..((((((....(.(((.((((((((((...((((....))))))))))))))))))))))))..))....(((((....)))))........))).))......... ( -41.00) >DroSim_CAF1 16315 120 + 1 CCGUGGACCAGGAACGCCUCCUCCGAUGGCGUAAAAAACGAAUAGCCCGUGGGCUUGUUUUUUACCGACGAGGCGAGCCAACAUGUGGUAACCCAUAAUUUACGCCUAGGGAUGUUAUGA ((.(((....((..((((((.((.....(.((((((((((...((((....))))))))))))))))).))))))..))....(((((....)))))........))).))......... ( -38.10) >consensus CCGUGGACCAGGAACGCCUCCUCCGAUGGCGUAAAAAACGAAUAGCCCGUGGGCUUGUUUUUUACCCACGAGGCGAGCCAACAUGUGGUAACCCAUAAUUUACGCCUAGGGAUGUUAUGA ((.(((....((..((((((....(.(((.((((((((((...((((....))))))))))))))))))))))))..))....(((((....)))))........))).))......... (-38.55 = -38.67 + 0.11)

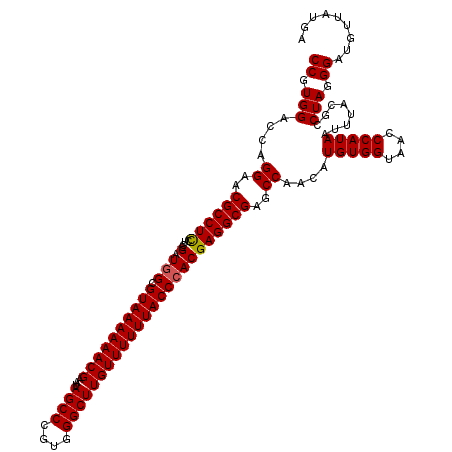

| Location | 21,511,618 – 21,511,738 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -40.70 |
| Consensus MFE | -40.50 |
| Energy contribution | -40.83 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21511618 120 - 23771897 UCAUAACAUCCCUAGGCGUAAAUUAUGGGUUACCACAUGUUGGCUCGCCUCGUGGGUAAAAAACAAGCCCACGGGCUAUUCGUUUUUUACGCCAUCGGAGAAGGCGUUCCUGGUCCACGG ..........((..((((((((..((((((..(((.....)))...(((.(((((((.........)))))))))).))))))..))))))))...(((..(((....)))..)))..)) ( -42.90) >DroSec_CAF1 16688 120 - 1 UCAUAACAUCCCUAGGCGUAAAUUAUGGGUUACCACAUGUUGGCUCGCCUCGUGGGUAAAAAACAAGCCCACGGGCUAUUCGUUUUUUACGCCAUCGGAGGAGGCGUUCCUGGUCCACGG ..........((..((((((((..((((((..(((.....)))...(((.(((((((.........)))))))))).))))))..))))))))...(((..(((....)))..)))..)) ( -42.50) >DroSim_CAF1 16315 120 - 1 UCAUAACAUCCCUAGGCGUAAAUUAUGGGUUACCACAUGUUGGCUCGCCUCGUCGGUAAAAAACAAGCCCACGGGCUAUUCGUUUUUUACGCCAUCGGAGGAGGCGUUCCUGGUCCACGG ..........((..((((((((..((((((((((..(((..((....)).))).)))))......((((....)))).)))))..))))))))...(((..(((....)))..)))..)) ( -36.70) >consensus UCAUAACAUCCCUAGGCGUAAAUUAUGGGUUACCACAUGUUGGCUCGCCUCGUGGGUAAAAAACAAGCCCACGGGCUAUUCGUUUUUUACGCCAUCGGAGGAGGCGUUCCUGGUCCACGG ..........((..((((((((..((((((..(((.....)))...(((.(((((((.........)))))))))).))))))..))))))))...(((..(((....)))..)))..)) (-40.50 = -40.83 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:08:07 2006