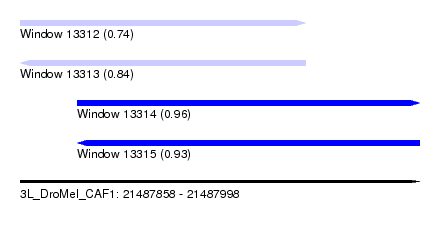
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,487,858 – 21,487,998 |
| Length | 140 |
| Max. P | 0.962059 |

| Location | 21,487,858 – 21,487,958 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.82 |
| Mean single sequence MFE | -33.82 |
| Consensus MFE | -27.18 |
| Energy contribution | -26.98 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.743135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21487858 100 + 23771897 -------------A-----A--CUCACCUUUGAGGUUGCGUGGUGCAGGAGGUAGUACCAUCUUCGCUGAUGGCGCUUCCCUUCCGAGCACGGUUCGCAGGUACGACCUCUUGUUGUCGU -------------.-----.--((((....)))).((((((.((((.(((((.((..(((((......)))))..))...)))))..)))).)..)))))..(((((........))))) ( -35.70) >DroVir_CAF1 16506 102 + 1 -------------A-----GCUCUCACCUUUGAGGUUGCGUGGUACUGGAAGUAGUACCAUCUUCGCUGAUGGUGCUUUCCUUCCGAAUACUGUUUGUGGGUACGACCUCUCUAUGUCGU -------------.-----(((((((....)))))..))((.((((((((((.(((((((((......)))))))))...)))))...(((.....)))))))).))............. ( -34.00) >DroGri_CAF1 10290 100 + 1 -------------A-----U--CUCACCUUUGAGGUUGCGUGGUACUGGAAGAAGUACCAUCUUCGCUGAUGGUGCUUUCCUUCCGAAUACUGUUUGUGGGUACGACCUCUCUAUGUCGU -------------(-----(--((((....))))))...((.((((((((((((((((((((......))))))))))..)))))...(((.....)))))))).))............. ( -33.30) >DroWil_CAF1 59688 102 + 1 ----------------UGAG--UUUACCUUUGAGGCUGCGUGGUGCAAGAAAUAAUACCAUCUUCGCUGAUGGUGCUUUCCUUCCGAGCCCUGUUUGUCGGUACGACCUCUCGUUGUCGU ----------------....--.........((((...((((.((((((.......((((((......))))))((((.......))))....)))).)).)))).)))).......... ( -23.80) >DroMoj_CAF1 10355 100 + 1 -------------A-----GC--CUACCUUUGAGGUUGCGUGGUACUGGAAGUAGUACCAUCUUCGCUGAUGGUGCUUUCCUUCCGAAUACUGUUUGUGGGUACGACCUCUCUAUGUCGU -------------.-----((--..(((.....))).))((.((((((((((.(((((((((......)))))))))...)))))...(((.....)))))))).))............. ( -34.10) >DroAna_CAF1 9097 118 + 1 CUACCAGUGCUACCAGUGAU--GUUACCUUUGAGGUUGCGUGGUGCAGGAGGUAGUACCAUCUUCGCUGAUGGCGCUUCCCUUCCGAGCACGGUUCGAAGGUACGACCUCUUGUUGUCGU .....(((((((.((((((.--((.(((.....))).))(((((((........)))))))..)))))).)))))))..(((((.((((...))))))))).(((((........))))) ( -42.00) >consensus _____________A_____G__CUCACCUUUGAGGUUGCGUGGUACAGGAAGUAGUACCAUCUUCGCUGAUGGUGCUUUCCUUCCGAACACUGUUUGUGGGUACGACCUCUCGAUGUCGU ...............................((((((.....((((.(((((.(((((((((......)))))))))...)))))((((...))))....)))))))))).......... (-27.18 = -26.98 + -0.19)

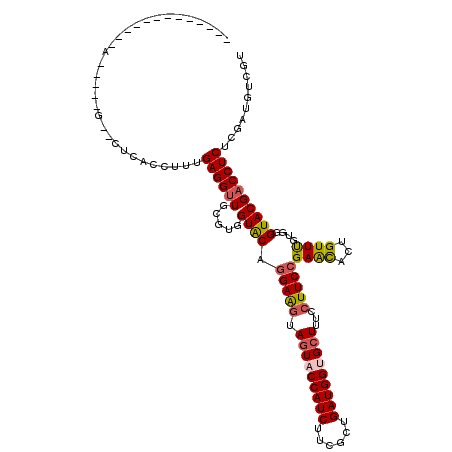

| Location | 21,487,858 – 21,487,958 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.82 |
| Mean single sequence MFE | -29.92 |
| Consensus MFE | -24.21 |
| Energy contribution | -23.18 |
| Covariance contribution | -1.03 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.844629 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21487858 100 - 23771897 ACGACAACAAGAGGUCGUACCUGCGAACCGUGCUCGGAAGGGAAGCGCCAUCAGCGAAGAUGGUACUACCUCCUGCACCACGCAACCUCAAAGGUGAG--U-----U------------- (((((........)))))...((((....((((..(((.((..((..(((((......)))))..)).))))).))))..))))..((((....))))--.-----.------------- ( -33.20) >DroVir_CAF1 16506 102 - 1 ACGACAUAGAGAGGUCGUACCCACAAACAGUAUUCGGAAGGAAAGCACCAUCAGCGAAGAUGGUACUACUUCCAGUACCACGCAACCUCAAAGGUGAGAGC-----U------------- (((((........)))))...........(((((.(((((...((.((((((......)))))).)).))))))))))...((.(((.....)))....))-----.------------- ( -28.20) >DroGri_CAF1 10290 100 - 1 ACGACAUAGAGAGGUCGUACCCACAAACAGUAUUCGGAAGGAAAGCACCAUCAGCGAAGAUGGUACUUCUUCCAGUACCACGCAACCUCAAAGGUGAG--A-----U------------- (((((........)))))...........(((((.((((((..((.((((((......)))))).)))))))))))))........((((....))))--.-----.------------- ( -27.30) >DroWil_CAF1 59688 102 - 1 ACGACAACGAGAGGUCGUACCGACAAACAGGGCUCGGAAGGAAAGCACCAUCAGCGAAGAUGGUAUUAUUUCUUGCACCACGCAGCCUCAAAGGUAAA--CUCA---------------- ..........(((....((((........(((((((..((((((..((((((......))))))....))))))......)).)))))....))))..--))).---------------- ( -27.30) >DroMoj_CAF1 10355 100 - 1 ACGACAUAGAGAGGUCGUACCCACAAACAGUAUUCGGAAGGAAAGCACCAUCAGCGAAGAUGGUACUACUUCCAGUACCACGCAACCUCAAAGGUAG--GC-----U------------- (((((........)))))...........(((((.(((((...((.((((((......)))))).)).))))))))))...((.(((.....)))..--))-----.------------- ( -28.70) >DroAna_CAF1 9097 118 - 1 ACGACAACAAGAGGUCGUACCUUCGAACCGUGCUCGGAAGGGAAGCGCCAUCAGCGAAGAUGGUACUACCUCCUGCACCACGCAACCUCAAAGGUAAC--AUCACUGGUAGCACUGGUAG (((((........)))))........((((((((((((.((..((..(((((......)))))..)).)))))(((.....)))(((.....)))...--.......).))))).))).. ( -34.80) >consensus ACGACAAAGAGAGGUCGUACCCACAAACAGUACUCGGAAGGAAAGCACCAUCAGCGAAGAUGGUACUACUUCCAGCACCACGCAACCUCAAAGGUAAG__C_____U_____________ (((((........)))))...........((((..(((((...((.((((((......)))))).)).))))).))))......(((.....)))......................... (-24.21 = -23.18 + -1.03)


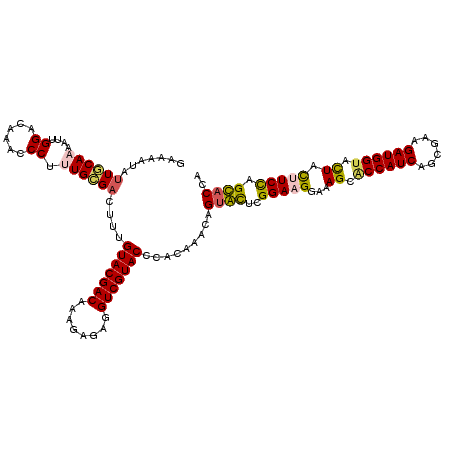
| Location | 21,487,878 – 21,487,998 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.94 |
| Mean single sequence MFE | -41.57 |
| Consensus MFE | -34.14 |
| Energy contribution | -35.17 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.962059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21487878 120 + 23771897 UGGUGCAGGAGGUAGUACCAUCUUCGCUGAUGGCGCUUCCCUUCCGAGCACGGUUCGCAGGUACGACCUCUUGUUGUCGUACGAAGUCGCAGAGGGUUUGUCCAAUUUUGCAAGAUUUUU ..((((.(((((.((..(((((......)))))..))...)))))..)))).........(((((((........)))))))(((((((((((.((.....))...)))))..)))))). ( -41.20) >DroVir_CAF1 16528 120 + 1 UGGUACUGGAAGUAGUACCAUCUUCGCUGAUGGUGCUUUCCUUCCGAAUACUGUUUGUGGGUACGACCUCUCUAUGUCGUACAAAGUCGCAAAGGGUUUGUCCAAUUUUGUAAAAUUUUC (((.((((((((.(((((((((......)))))))))...))))))...(((.((((((((((((((........)))))))....))))))).)))..)))))................ ( -41.80) >DroPse_CAF1 16195 120 + 1 UGGUGCAGGAGGAAGUACCAUCUUCGCUGAUGGAGCUUCCCUUCCGAGCACUGUUUGGGGGUACGACCUCUUGUUGUCGUACAAAGUCACAGAGGGUUUGUCCAAUUUUGUAUGAUUUUC .(((((.(((((((((.(((((......))))).))))))..)))..)))))....((((......)))).....((((((((((((.(((((...)))))...)))))))))))).... ( -44.00) >DroGri_CAF1 10310 120 + 1 UGGUACUGGAAGAAGUACCAUCUUCGCUGAUGGUGCUUUCCUUCCGAAUACUGUUUGUGGGUACGACCUCUCUAUGUCGUACAAAGUCGCAAAGGGUUUGUCCAAUUUUGCAAUAUUUUC (((.((((((((((((((((((......))))))))))..))))))...(((.((((((((((((((........)))))))....))))))).)))..)))))................ ( -42.10) >DroWil_CAF1 59710 120 + 1 UGGUGCAAGAAAUAAUACCAUCUUCGCUGAUGGUGCUUUCCUUCCGAGCCCUGUUUGUCGGUACGACCUCUCGUUGUCGUACAAAGUCGCAAAGGGUUUGUCCAAUUUUGCAAACUUUUU ...(((((((.....(((((((......))))))).........((((((((...((.(.(((((((........)))))))...).))...)))))))).....)))))))........ ( -38.50) >DroMoj_CAF1 10375 120 + 1 UGGUACUGGAAGUAGUACCAUCUUCGCUGAUGGUGCUUUCCUUCCGAAUACUGUUUGUGGGUACGACCUCUCUAUGUCGUACAAAGUCGCAAAGGGUUUGUCCAAUUUUGCAAUAUUUUC (((.((((((((.(((((((((......)))))))))...))))))...(((.((((((((((((((........)))))))....))))))).)))..)))))................ ( -41.80) >consensus UGGUACAGGAAGUAGUACCAUCUUCGCUGAUGGUGCUUUCCUUCCGAACACUGUUUGUGGGUACGACCUCUCGAUGUCGUACAAAGUCGCAAAGGGUUUGUCCAAUUUUGCAAAAUUUUC (((.((.(((((.(((((((((......)))))))))...)))))....(((.((((((((((((((........)))))))....))))))).)))..)))))................ (-34.14 = -35.17 + 1.03)



| Location | 21,487,878 – 21,487,998 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.94 |
| Mean single sequence MFE | -34.20 |
| Consensus MFE | -28.75 |
| Energy contribution | -28.58 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925124 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21487878 120 - 23771897 AAAAAUCUUGCAAAAUUGGACAAACCCUCUGCGACUUCGUACGACAACAAGAGGUCGUACCUGCGAACCGUGCUCGGAAGGGAAGCGCCAUCAGCGAAGAUGGUACUACCUCCUGCACCA .......(((((.....((......))..(((((((((............)))))))))..)))))...((((..(((.((..((..(((((......)))))..)).))))).)))).. ( -37.80) >DroVir_CAF1 16528 120 - 1 GAAAAUUUUACAAAAUUGGACAAACCCUUUGCGACUUUGUACGACAUAGAGAGGUCGUACCCACAAACAGUAUUCGGAAGGAAAGCACCAUCAGCGAAGAUGGUACUACUUCCAGUACCA ................(((.(((.....))).......(((((((........))))))))))......(((((.(((((...((.((((((......)))))).)).)))))))))).. ( -29.70) >DroPse_CAF1 16195 120 - 1 GAAAAUCAUACAAAAUUGGACAAACCCUCUGUGACUUUGUACGACAACAAGAGGUCGUACCCCCAAACAGUGCUCGGAAGGGAAGCUCCAUCAGCGAAGAUGGUACUUCCUCCUGCACCA ...............(((((((.......)))......(((((((........)))))))..))))...((((..(((..(((((..(((((......)))))..)))))))).)))).. ( -35.40) >DroGri_CAF1 10310 120 - 1 GAAAAUAUUGCAAAAUUGGACAAACCCUUUGCGACUUUGUACGACAUAGAGAGGUCGUACCCACAAACAGUAUUCGGAAGGAAAGCACCAUCAGCGAAGAUGGUACUUCUUCCAGUACCA .......(((((((...((......)))))))))....(((((((........))))))).........(((((.((((((..((.((((((......)))))).))))))))))))).. ( -34.70) >DroWil_CAF1 59710 120 - 1 AAAAAGUUUGCAAAAUUGGACAAACCCUUUGCGACUUUGUACGACAACGAGAGGUCGUACCGACAAACAGGGCUCGGAAGGAAAGCACCAUCAGCGAAGAUGGUAUUAUUUCUUGCACCA ..((((((.(((((...((......)))))))))))))(((((((........))))))).........((((..(((((..((..((((((......)))))).)).))))).)).)). ( -32.90) >DroMoj_CAF1 10375 120 - 1 GAAAAUAUUGCAAAAUUGGACAAACCCUUUGCGACUUUGUACGACAUAGAGAGGUCGUACCCACAAACAGUAUUCGGAAGGAAAGCACCAUCAGCGAAGAUGGUACUACUUCCAGUACCA .......(((((((...((......)))))))))....(((((((........))))))).........(((((.(((((...((.((((((......)))))).)).)))))))))).. ( -34.70) >consensus GAAAAUAUUGCAAAAUUGGACAAACCCUUUGCGACUUUGUACGACAAAGAGAGGUCGUACCCACAAACAGUACUCGGAAGGAAAGCACCAUCAGCGAAGAUGGUACUACUUCCAGCACCA .......((((((....((......)).))))))....(((((((........))))))).........((((..(((((...((.((((((......)))))).)).))))).)))).. (-28.75 = -28.58 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:07:35 2006