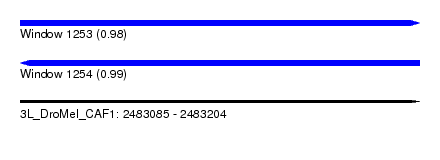
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,483,085 – 2,483,204 |
| Length | 119 |
| Max. P | 0.988166 |

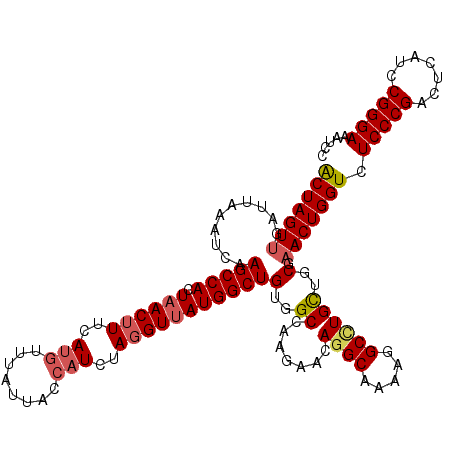
| Location | 2,483,085 – 2,483,204 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 93.36 |
| Mean single sequence MFE | -36.00 |
| Consensus MFE | -35.24 |
| Energy contribution | -35.20 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2483085 119 + 23771897 GGAUUUCCCGGAUGAGUCGGGAGACCAGUUCGCAGCAGGCCUUUUGCCUGUUCUUGGCCACAGCCAUAACCUAGAUGGUGAUAAACAUGAAAGUUAGUGGCUUAAUUUAAUCAACUAGU ((.((((((((.....))))))))))((((.(.(((((((.....))))))))..((((((.(((((.......)))))....(((......))).))))))..........))))... ( -37.30) >DroSec_CAF1 106580 119 + 1 GGAUUUCCCGGAUGAGUCGGGAGACCAGUUCGCAGCAAGCCUUUUGCCUGUUCUUGGCCACAGCCAUUACCUAGAUGGUAAUAAACAUGAAAGUUAGUGGCUGGAUUUAAUCAACUAGU ((.((((((((.....))))))))))((((.((((((((...)))).))))..(..(((((.((((((.....))))))....(((......))).)))))..)........))))... ( -35.10) >DroSim_CAF1 101491 119 + 1 GGAUUUCCCGGAUGAGUCGGGAGACCAGUUCGCAGCAGGCCUUUUGCCUGUUCUUGGCCACAGCCAUAACCUAGAUGGUAAUAAAUUUGAAAGUUAGUGGCUUGAUUUAAUUAACUAGU ((.((((((((.....))))))))))((((....((((((.....))))))((..((((((.(((((.......)))))....((((....)))).)))))).)).......))))... ( -35.70) >DroEre_CAF1 106813 119 + 1 GGAUCUCCCGGAUAAGUCGGGAGACCAGUUCGCAGCAGGCCUUUUGCCUGUUCUUGGCCACAGCCAUAACCUAGAUGGUGAUAAGCAUUAAAGUUAGUGGCUUGAUUUAAUUCACUAGC ((.((((((((.....)))))))))).....(((((((((.....)))))))..((((....))))..(((.....))).....))......(((((((..(((...)))..))))))) ( -40.40) >DroYak_CAF1 104727 119 + 1 GGAUUUCCCGGAUGAGUCGGGAGACCAGUUCGCAACAUGCCUUCUGCCUGUUCUUGGCCACAGCCAUAACCUAGAUGGUAAUAAACAUGAAUGUUAGUGGCUUGAUUAAAUCAACUAGU ((.((((((((.....))))))))))((((.(.((((.((.....)).)))))..((((((.(((((.......)))))....((((....)))).))))))..........))))... ( -31.50) >consensus GGAUUUCCCGGAUGAGUCGGGAGACCAGUUCGCAGCAGGCCUUUUGCCUGUUCUUGGCCACAGCCAUAACCUAGAUGGUAAUAAACAUGAAAGUUAGUGGCUUGAUUUAAUCAACUAGU ((.((((((((.....))))))))))((((.(.(((((((.....))))))))..((((((.(((((.......)))))....(((......))).))))))..........))))... (-35.24 = -35.20 + -0.04)

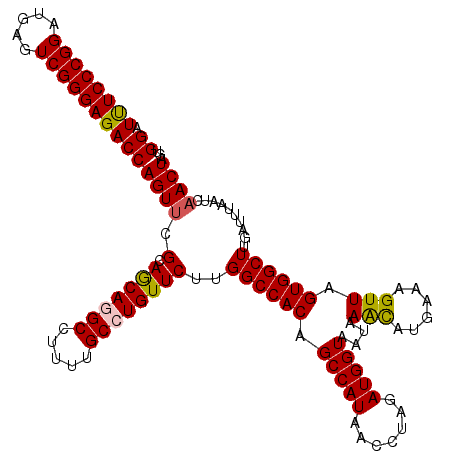
| Location | 2,483,085 – 2,483,204 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 93.36 |
| Mean single sequence MFE | -33.42 |
| Consensus MFE | -30.21 |
| Energy contribution | -30.97 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988166 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2483085 119 - 23771897 ACUAGUUGAUUAAAUUAAGCCACUAACUUUCAUGUUUAUCACCAUCUAGGUUAUGGCUGUGGCCAAGAACAGGCAAAAGGCCUGCUGCGAACUGGUCUCCCGACUCAUCCGGGAAAUCC (((((((..........(((((.((((((..(((........)))..)))))))))))(..((.......((((.....))))))..).))))))).(((((.......)))))..... ( -35.01) >DroSec_CAF1 106580 119 - 1 ACUAGUUGAUUAAAUCCAGCCACUAACUUUCAUGUUUAUUACCAUCUAGGUAAUGGCUGUGGCCAAGAACAGGCAAAAGGCUUGCUGCGAACUGGUCUCCCGACUCAUCCGGGAAAUCC (((((((...........(((((.(((......)))(((((((.....)))))))...)))))......(((((.....))))).....))))))).(((((.......)))))..... ( -32.70) >DroSim_CAF1 101491 119 - 1 ACUAGUUAAUUAAAUCAAGCCACUAACUUUCAAAUUUAUUACCAUCUAGGUUAUGGCUGUGGCCAAGAACAGGCAAAAGGCCUGCUGCGAACUGGUCUCCCGACUCAUCCGGGAAAUCC (((((((..........(((((.((((((..................)))))))))))(..((.......((((.....))))))..).))))))).(((((.......)))))..... ( -31.98) >DroEre_CAF1 106813 119 - 1 GCUAGUGAAUUAAAUCAAGCCACUAACUUUAAUGCUUAUCACCAUCUAGGUUAUGGCUGUGGCCAAGAACAGGCAAAAGGCCUGCUGCGAACUGGUCUCCCGACUUAUCCGGGAGAUCC ((((.(((......)))(((((.((((((..(((........)))..))))))))))).))))......(((((.....))))).........(((((((((.......))))))))). ( -38.50) >DroYak_CAF1 104727 119 - 1 ACUAGUUGAUUUAAUCAAGCCACUAACAUUCAUGUUUAUUACCAUCUAGGUUAUGGCUGUGGCCAAGAACAGGCAGAAGGCAUGUUGCGAACUGGUCUCCCGACUCAUCCGGGAAAUCC (((((((.......((.(((((.((((....(((........)))....)))))))))...(((.......))).))..((.....)).))))))).(((((.......)))))..... ( -28.90) >consensus ACUAGUUGAUUAAAUCAAGCCACUAACUUUCAUGUUUAUUACCAUCUAGGUUAUGGCUGUGGCCAAGAACAGGCAAAAGGCCUGCUGCGAACUGGUCUCCCGACUCAUCCGGGAAAUCC (((((((..........(((((.((((((..(((........)))..)))))))))))(..((.......((((.....))))))..).))))))).(((((.......)))))..... (-30.21 = -30.97 + 0.76)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:53 2006