
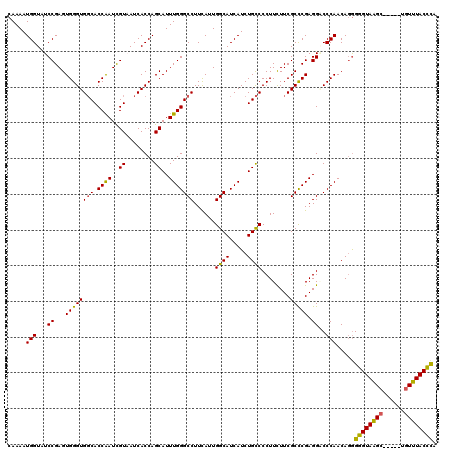
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,473,824 – 21,474,014 |
| Length | 190 |
| Max. P | 0.956661 |

| Location | 21,473,824 – 21,473,944 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.65 |
| Mean single sequence MFE | -38.42 |
| Consensus MFE | -34.75 |
| Energy contribution | -34.25 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21473824 120 + 23771897 CAAAAUGGUAUCCGAGUGGGUGGCACCAAUCGUUAUCACCAGCAUUUGGGCCUUCAUUGGCAUCAUCUGCCCCUUCUUCGCCCGAGGACCCAACAGGGGGUAAGCUAAUCUGUUUACCCA ...........((...(((((((((((((..(((......)))..))))(((......)))......))))....((((....)))))))))...))((((((((......)))))))). ( -41.10) >DroSec_CAF1 10724 120 + 1 CAAAAUGGUAUCCGAGUGGGUGGCACCAAUUGUAAUCACCAGCAUUUGGGCCUUCAUUGGCAUCAUCUGCCCCUUUUUCGCCCGAGGACCCAACAGGGGGUAAGCAAAUCUGUUUACCCA ...........((...(((((((((((((.(((........))).))))(((......)))......))))((((........)))))))))...))(((((((((....))))))))). ( -43.00) >DroSim_CAF1 9235 120 + 1 CAAAAUGGUAUCCGAGUGGGUGGCACCAAUUGUAAUCACCAGCAUUUGGGCCUUUAUUGGCAUCAUCUGCCCCUUUUUCGCCCGAGGACCCAACAGGGGGUAAGCAAAUCUGUUUACCCA ...........((...(((((((((((((.(((........))).))))(((......)))......))))((((........)))))))))...))(((((((((....))))))))). ( -43.00) >DroEre_CAF1 10831 115 + 1 CAAAAUGGUUUCCGAGUGGGUGGCACCAAUCGUAAUCACCAGCAUUUGGGCCUUCAUUGGCAUCAUCUGCCCCUUCUUCGCCCGAGGGCCCAACAGAGGGUAAGC-----UGUUUACCCA .....((((((.(((.(((......))).))).))..))))....((((((((((...((((.....)))).(......)...))))))))))....(((((((.-----..))))))). ( -37.00) >DroYak_CAF1 19750 115 + 1 CAAAAUGGUUUCCGAGUGGGUGGCACCAAUCGUAAUCACCAGCAUUUGGGCCUUCAUUGGCAUCAUCUGCCCCUUCUUCGCCCGAGGUCCCAACAGGGGGUAAGC-----UGUUUACCCA .....(((...((...((((((((.((((..((........))..)))))))......((((.....))))........))))).))..))).....(((((((.-----..))))))). ( -34.10) >DroAna_CAF1 9542 114 + 1 CAAAAUGGUUUCCGAGUGGGUGCCACCGAUCGUUAUCACCAGCAUUUGGGCCUUUAUUGGCAUCAUCUGUCCCUUCUUCGCUCGAGGUCCCAACAAGGGGUAGG------UGUUUACUUA .....((((.(((....))).))))((((..(((......)))..))))(((......)))..(((((..(((((((((....)))).......)))))..)))------))........ ( -32.31) >consensus CAAAAUGGUAUCCGAGUGGGUGGCACCAAUCGUAAUCACCAGCAUUUGGGCCUUCAUUGGCAUCAUCUGCCCCUUCUUCGCCCGAGGACCCAACAGGGGGUAAGC_____UGUUUACCCA .....(((...((...((((((((.((((..((........))..)))))))......((((.....))))........))))).))..))).....((((((((......)))))))). (-34.75 = -34.25 + -0.50)


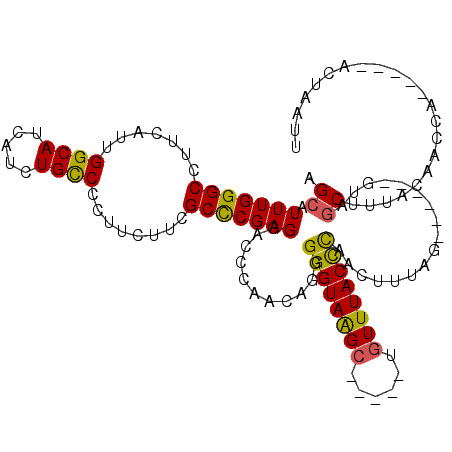
| Location | 21,473,864 – 21,473,979 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.41 |
| Mean single sequence MFE | -29.41 |
| Consensus MFE | -24.43 |
| Energy contribution | -24.08 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21473864 115 + 23771897 AGCAUUUGGGCCUUCAUUGGCAUCAUCUGCCCCUUCUUCGCCCGAGGACCCAACAGGGGGUAAGCUAAUCUGUUUACCCAACUUCAGUCUUGAGUAGCUUUGCAACCC-----ACAAAUG .(((..(((((.......((((.....))))........)))))(((((((....))((((((((......)))))))).......))))).........))).....-----....... ( -32.56) >DroSec_CAF1 10764 115 + 1 AGCAUUUGGGCCUUCAUUGGCAUCAUCUGCCCCUUUUUCGCCCGAGGACCCAACAGGGGGUAAGCAAAUCUGUUUACCCAACUUUAGUCUUGAGUAGCUUUGCAACCA-----ACUAAUU .(((..(((((.......((((.....))))........)))))(((((((....))(((((((((....))))))))).......))))).........))).....-----....... ( -32.96) >DroSim_CAF1 9275 115 + 1 AGCAUUUGGGCCUUUAUUGGCAUCAUCUGCCCCUUUUUCGCCCGAGGACCCAACAGGGGGUAAGCAAAUCUGUUUACCCAACUUUAGUCUUGAGUAGCUUUGCAACCA-----ACUAAUU .(((..(((((.......((((.....))))........)))))(((((((....))(((((((((....))))))))).......))))).........))).....-----....... ( -32.96) >DroEre_CAF1 10871 103 + 1 AGCAUUUGGGCCUUCAUUGGCAUCAUCUGCCCCUUCUUCGCCCGAGGGCCCAACAGAGGGUAAGC-----UGUUUACCCAACUUUAG-------UAGCUGUAGAACCA-----ACUAAUU (((..((((((((((...((((.....)))).(......)...))))))))))....(((((((.-----..)))))))........-------..))).........-----....... ( -30.30) >DroYak_CAF1 19790 108 + 1 AGCAUUUGGGCCUUCAUUGGCAUCAUCUGCCCCUUCUUCGCCCGAGGUCCCAACAGGGGGUAAGC-----UGUUUACCCAACUUUAG-------UUCCCUUACAACUAACUUAACUAAUU .((.(((((((.......((((.....))))........))))))))).((....))(((((((.-----..)))))))....((((-------((.......))))))........... ( -23.76) >DroAna_CAF1 9582 99 + 1 AGCAUUUGGGCCUUUAUUGGCAUCAUCUGUCCCUUCUUCGCUCGAGGUCCCAACAAGGGGUAGG------UGUUUACUUAAUAU---------GUUGCUU-AAACCCU-----CCCACUU ((((.....(((......)))..(((((..(((((((((....)))).......)))))..)))------))............---------..)))).-.......-----....... ( -23.91) >consensus AGCAUUUGGGCCUUCAUUGGCAUCAUCUGCCCCUUCUUCGCCCGAGGACCCAACAGGGGGUAAGC_____UGUUUACCCAACUUUAG______GUAGCUUUACAACCA_____ACUAAUU .((.(((((((.......((((.....))))........)))))))...........((((((((......)))))))).................))...................... (-24.43 = -24.08 + -0.36)

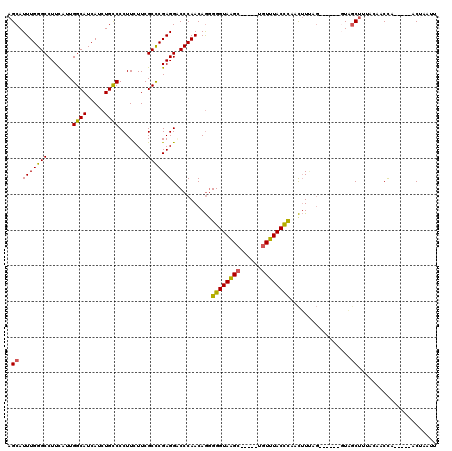
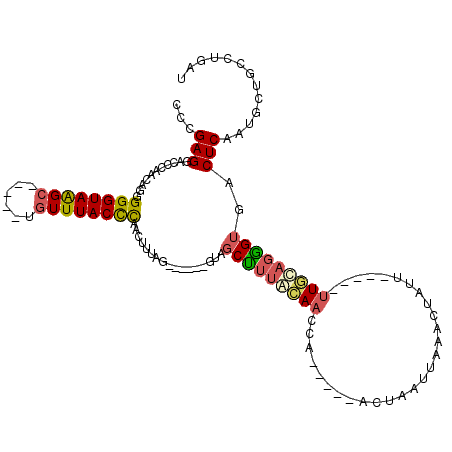
| Location | 21,473,904 – 21,474,014 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.85 |
| Mean single sequence MFE | -29.81 |
| Consensus MFE | -16.85 |
| Energy contribution | -17.11 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21473904 110 + 23771897 CCCGAGGACCCAACAGGGGGUAAGCUAAUCUGUUUACCCAACUUCAGUCUUGAGUAGCUUUGCAACCC-----ACAAAUGAAACUAUU-----UUGCAGGGUGACUCAAUGCUGCCUGAU .............((((((((((((......)))))))).....((((.((((((.(((((((((...-----...............-----))))))))).)))))).)))))))).. ( -39.17) >DroSec_CAF1 10804 110 + 1 CCCGAGGACCCAACAGGGGGUAAGCAAAUCUGUUUACCCAACUUUAGUCUUGAGUAGCUUUGCAACCA-----ACUAAUUAAACUAUU-----UUGCAGGGUGACUCAAUGCUGCCUGAU .............(((((((((((((....))))))))).....((((.((((((.(((((((((...-----...............-----))))))))).)))))).)))))))).. ( -38.17) >DroSim_CAF1 9315 110 + 1 CCCGAGGACCCAACAGGGGGUAAGCAAAUCUGUUUACCCAACUUUAGUCUUGAGUAGCUUUGCAACCA-----ACUAAUUAAACUAUU-----UUGCAGGGUGACUCAAUGCUGCCUGAU .............(((((((((((((....))))))))).....((((.((((((.(((((((((...-----...............-----))))))))).)))))).)))))))).. ( -38.17) >DroEre_CAF1 10911 98 + 1 CCCGAGGGCCCAACAGAGGGUAAGC-----UGUUUACCCAACUUUAG-------UAGCUGUAGAACCA-----ACUAAUUUAACUAUU-----UUGCAGAGUGACUCAAUGCUGCCUGAU ((....)).....(((.(((((((.-----..))))))).......(-------(((((((((((...-----.............))-----)))))(((...)))...)))))))).. ( -20.09) >DroYak_CAF1 19830 103 + 1 CCCGAGGUCCCAACAGGGGGUAAGC-----UGUUUACCCAACUUUAG-------UUCCCUUACAACUAACUUAACUAAUUUAAAAAUU-----UUGCAGGGUGACUCAAUGCUGCCUGAU .............((((.((((...-----.(((.((((....((((-------((.......)))))).((((.....)))).....-----.....)))))))....)))).)))).. ( -20.10) >DroAna_CAF1 9622 99 + 1 CUCGAGGUCCCAACAAGGGGUAGG------UGUUUACUUAAUAU---------GUUGCUU-AAACCCU-----CCCACUUAAUAUAUUCCCGAUUUUAGGGUGACUCAAUGCUGCCUUAU .(((.((.......((((((.(((------.(((((..(((...---------.)))..)-)))))))-----))).)))........)))))...((((((..(.....)..)))))). ( -23.16) >consensus CCCGAGGACCCAACAGGGGGUAAGC_____UGUUUACCCAACUUUAG______GUAGCUUUACAACCA_____ACUAAUUAAACUAUU_____UUGCAGGGUGACUCAAUGCUGCCUGAU ...(((...........((((((((......)))))))).................(((((((((............................)))))))))..)))............. (-16.85 = -17.11 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:07:22 2006