
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,445,468 – 21,445,561 |
| Length | 93 |
| Max. P | 0.800664 |

| Location | 21,445,468 – 21,445,561 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 81.29 |
| Mean single sequence MFE | -20.18 |
| Consensus MFE | -14.74 |
| Energy contribution | -14.02 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.800664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
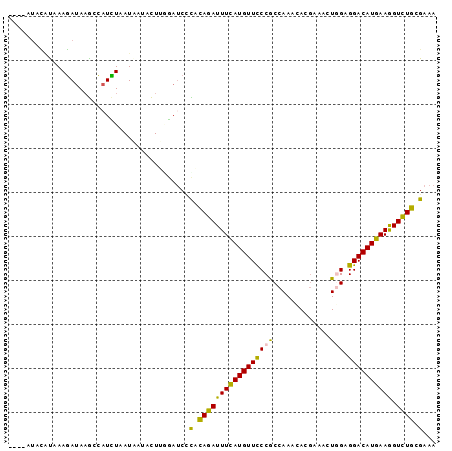
>3L_DroMel_CAF1 21445468 93 + 23771897 ----AUACAUAAAGAUAAGCCAUCUAAUACUACUUCGAUCCCACAGAUUUUAUGUUCCCGCCAAACACGAAACUGGAGGACAUGAAGGUCUGCGAAA ----........((((.....))))................(.((((((((((((((...(((..........))).))))))))).))))).)... ( -16.60) >DroSec_CAF1 4135 93 + 1 ----AUACAUAAAGAUAAGCCAUCUAAUAAUACUUGGAUCCCACAGAUUUCAUGUUCCCGCCAAACACGAAAUUGGAGGACAUGAAGGUCUGCGAAA ----.................((((((......))))))..(.((((((((((((((...((((........)))).))))))))).))))).)... ( -21.90) >DroSim_CAF1 1600 97 + 1 AGAGAUACAUAAAGAUAAGCCAUCUAAUAAUACUUGGAUCCCACAGAUUUCAUGUUCCCGCCAAACACGAAAUUGGAGGACAUGAAGGUCUGCGAAA .....................((((((......))))))..(.((((((((((((((...((((........)))).))))))))).))))).)... ( -21.90) >DroEre_CAF1 4180 92 + 1 ----AUACGCAAAGAUAAGUCUUUUAAUAAUUCUG-GAUCCCACAGAUUUCAUGUUUCCGCCAAACACGAAACUUGAGGACAUGAAGGUCUGCGAAA ----...((((..(((...............((((-.......))))(((((((((..((..............))..))))))))))))))))... ( -18.44) >DroWil_CAF1 24223 93 + 1 -AUCAAAAAUGAAUAUAAAUAAUAUAAUGUUAUUCAU---UUGUAGAUUUCAUGUUCCCACCAAAUACCAAACUGGAGGACAUGAAAGUUUGCGAGA -.((..(((((((((...((......))..)))))))---))((((((((((((((((((.............))).))))))).)))))))))).. ( -23.12) >DroYak_CAF1 1713 93 + 1 ----AUAUCCAAAGAUACGCCAUUUAAUAUUUCUGGGAUCGUACAGAUUUCAUGUUCCCGCCAAACACGAAACUUGAGGACAUGAAGGUCUGCGAAA ----...((((.(((((..........))))).)))).(((..((((((((((((((.((..............)).))))))))).)))))))).. ( -19.14) >consensus ____AUACAUAAAGAUAAGCCAUCUAAUAAUACUUGGAUCCCACAGAUUUCAUGUUCCCGCCAAACACGAAACUGGAGGACAUGAAGGUCUGCGAAA .........................................(.(((((((((((((((((.............))).)))))))))).)))).)... (-14.74 = -14.02 + -0.72)

| Location | 21,445,468 – 21,445,561 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 81.29 |
| Mean single sequence MFE | -23.16 |
| Consensus MFE | -16.59 |
| Energy contribution | -17.95 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
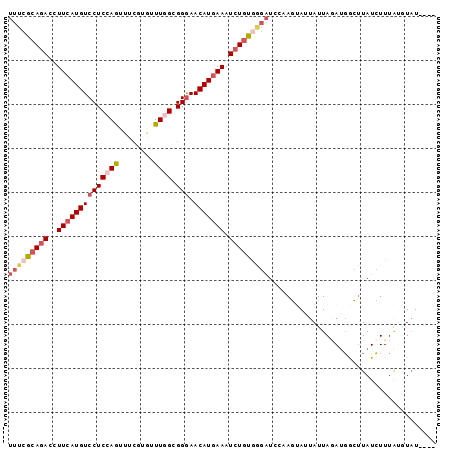
>3L_DroMel_CAF1 21445468 93 - 23771897 UUUCGCAGACCUUCAUGUCCUCCAGUUUCGUGUUUGGCGGGAACAUAAAAUCUGUGGGAUCGAAGUAGUAUUAGAUGGCUUAUCUUUAUGUAU---- (..((((((..((.(((((((((((........)))).))).)))).)).))))))..)..((((.(((........)))...))))......---- ( -21.10) >DroSec_CAF1 4135 93 - 1 UUUCGCAGACCUUCAUGUCCUCCAAUUUCGUGUUUGGCGGGAACAUGAAAUCUGUGGGAUCCAAGUAUUAUUAGAUGGCUUAUCUUUAUGUAU---- (..((((((..((((((((((((((........)))).))).))))))).))))))..).....((((....(((((...)))))....))))---- ( -27.20) >DroSim_CAF1 1600 97 - 1 UUUCGCAGACCUUCAUGUCCUCCAAUUUCGUGUUUGGCGGGAACAUGAAAUCUGUGGGAUCCAAGUAUUAUUAGAUGGCUUAUCUUUAUGUAUCUCU (..((((((..((((((((((((((........)))).))).))))))).))))))..).....((((....(((((...)))))....)))).... ( -27.30) >DroEre_CAF1 4180 92 - 1 UUUCGCAGACCUUCAUGUCCUCAAGUUUCGUGUUUGGCGGAAACAUGAAAUCUGUGGGAUC-CAGAAUUAUUAAAAGACUUAUCUUUGCGUAU---- ...(((.....(((..(((((((..((((((((((.....))))))))))..)).))))).-..)))......(((((....))))))))...---- ( -22.40) >DroWil_CAF1 24223 93 - 1 UCUCGCAAACUUUCAUGUCCUCCAGUUUGGUAUUUGGUGGGAACAUGAAAUCUACAA---AUGAAUAACAUUAUAUUAUUUAUAUUCAUUUUUGAU- ..........(((((((((((((((........)))).))).))))))))((...((---(((((((...............)))))))))..)).- ( -19.66) >DroYak_CAF1 1713 93 - 1 UUUCGCAGACCUUCAUGUCCUCAAGUUUCGUGUUUGGCGGGAACAUGAAAUCUGUACGAUCCCAGAAAUAUUAAAUGGCGUAUCUUUGGAUAU---- ....(((((..((((((((((((((.......))))..))).))))))).)))))......(((((.((((........)))).)))))....---- ( -21.30) >consensus UUUCGCAGACCUUCAUGUCCUCCAGUUUCGUGUUUGGCGGGAACAUGAAAUCUGUGGGAUCCAAGUAUUAUUAGAUGGCUUAUCUUUAUGUAU____ (((((((((..((((((((((((((........)))).))).))))))).)))))))))...................................... (-16.59 = -17.95 + 1.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:07:12 2006