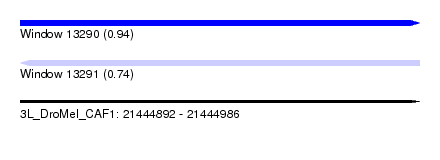
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,444,892 – 21,444,986 |
| Length | 94 |
| Max. P | 0.935003 |

| Location | 21,444,892 – 21,444,986 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 77.32 |
| Mean single sequence MFE | -21.57 |
| Consensus MFE | -15.65 |
| Energy contribution | -15.73 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21444892 94 + 23771897 AAAGUUUAUUGAUAUUAUUUUUCCUAAAA-A------------UGUUUUUAGGAGAAUGCUGCCAAUCCUGCAUGGCGCGUAUUCCCUACGCCACCCUGAUAGCCAC ..................(((((((((((-.------------...))))))))))).(((((((........))))(((((.....))))).........)))... ( -22.00) >DroSec_CAF1 3540 105 + 1 AAAGUUUAUUGAAAACAUAUUUCCUAAAA-U-GUGCCACAUUUUGUUUUUAGGAGAAUGCUGCCAAUCCUGCAUGGCGCGUAUUCCCUACGCCACCCUGAUAGCCAC ...(.(((((..............(((((-(-(.....)))))))....((((.(((((((((((........))))).)))))))))).........))))).).. ( -21.80) >DroSim_CAF1 1005 105 + 1 AAAGUUUAUUGAAAACAUAUUUUCUAAAA-U-GUGCCACAUUUUGUUUUUAGGAGAAUGCUGCCAAUCCUGCAUGGCGCGUAUUCCCUACGCCACCCUGAUAGCCAC ...(.((((((((((....)))))(((((-(-(.....)))))))....((((.(((((((((((........))))).)))))))))).........))))).).. ( -22.20) >DroEre_CAF1 3593 101 + 1 AAACUU-UUUAAAAAUAUUUUUCCUCAAA-U-GUAACUCAUU---UUUGUAGGAGAAUGCUGCCAAUCCUGCAUGGCGCGUAUUCCCUACGCCACCCUGAUAGCCAC ......-...................(((-(-(.....))))---)..(((((.(((((((((((........))))).)))))))))))................. ( -22.00) >DroYak_CAF1 1127 101 + 1 AAAGUU-AUUGAUAAACUUCUUCCUAAAA-U-GUAACUCGUU---UCUUUAGGAGAAUGCUGCCAAUCCUGCAUGGCGCGUAUUCCCUACGCCACCCUGAUAGCCAC ...(((-(((...........((((((((-(-(.....))).---..)))))))(((((((((((........))))).)))))).............))))))... ( -21.60) >DroMoj_CAF1 2733 88 + 1 ----UUUAUUGUGAAUUAAUU--CUA--AUUUU-----------AAUUUUAGCGGACUGUUGCCAAUCAUGUAUGGCACGCAUACCCUAUGCAACGCUAAUAGCAAC ----....(((((((((((..--...--...))-----------)))))(((((......(((((........))))).(((((...)))))..)))))...)))). ( -19.80) >consensus AAAGUUUAUUGAAAAUAUAUUUCCUAAAA_U_GU__C_CAUU__GUUUUUAGGAGAAUGCUGCCAAUCCUGCAUGGCGCGUAUUCCCUACGCCACCCUGAUAGCCAC .................................................((((.(((((((((((........))))).)))))))))).................. (-15.65 = -15.73 + 0.08)


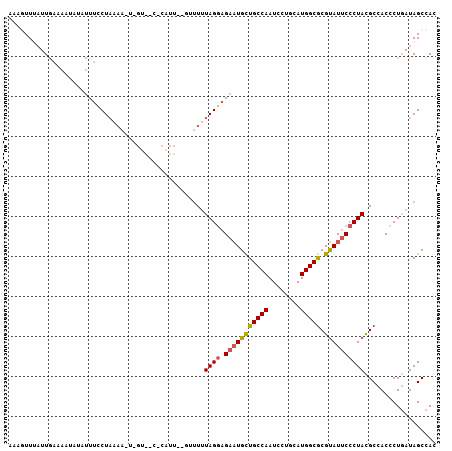
| Location | 21,444,892 – 21,444,986 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 77.32 |
| Mean single sequence MFE | -24.33 |
| Consensus MFE | -17.45 |
| Energy contribution | -18.10 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.739950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21444892 94 - 23771897 GUGGCUAUCAGGGUGGCGUAGGGAAUACGCGCCAUGCAGGAUUGGCAGCAUUCUCCUAAAAACA------------U-UUUUAGGAAAAAUAAUAUCAAUAAACUUU ((.((((((.(.(((((((.(......)))))))).)..)).)))).))....((((((((...------------.-))))))))..................... ( -25.30) >DroSec_CAF1 3540 105 - 1 GUGGCUAUCAGGGUGGCGUAGGGAAUACGCGCCAUGCAGGAUUGGCAGCAUUCUCCUAAAAACAAAAUGUGGCAC-A-UUUUAGGAAAUAUGUUUUCAAUAAACUUU ((((((((....)))(((((.....))))))))))..((.(((((.(((((..((((((((..............-.-))))))))...))))).)))))...)).. ( -25.86) >DroSim_CAF1 1005 105 - 1 GUGGCUAUCAGGGUGGCGUAGGGAAUACGCGCCAUGCAGGAUUGGCAGCAUUCUCCUAAAAACAAAAUGUGGCAC-A-UUUUAGAAAAUAUGUUUUCAAUAAACUUU (((((((.((...((...((((((((..((((((........)))).)))))).))))....))...)))))).)-)-)....(((((....))))).......... ( -24.90) >DroEre_CAF1 3593 101 - 1 GUGGCUAUCAGGGUGGCGUAGGGAAUACGCGCCAUGCAGGAUUGGCAGCAUUCUCCUACAAA---AAUGAGUUAC-A-UUUGAGGAAAAAUAUUUUUAAA-AAGUUU ((((((((....)))))(((((((((..((((((........)))).)))))).)))))...---.......)))-.-((((((((......))))))))-...... ( -26.30) >DroYak_CAF1 1127 101 - 1 GUGGCUAUCAGGGUGGCGUAGGGAAUACGCGCCAUGCAGGAUUGGCAGCAUUCUCCUAAAGA---AACGAGUUAC-A-UUUUAGGAAGAAGUUUAUCAAU-AACUUU ((.((((((.(.(((((((.(......)))))))).)..)).)))).))....((((((((.---..........-.-)))))))).((((((.......-)))))) ( -25.50) >DroMoj_CAF1 2733 88 - 1 GUUGCUAUUAGCGUUGCAUAGGGUAUGCGUGCCAUACAUGAUUGGCAACAGUCCGCUAAAAUU-----------AAAAU--UAG--AAUUAAUUCACAAUAAA---- ((((...((((((..(((((...))))).(((((........)))))......))))))....-----------.....--..(--((....))).))))...---- ( -18.10) >consensus GUGGCUAUCAGGGUGGCGUAGGGAAUACGCGCCAUGCAGGAUUGGCAGCAUUCUCCUAAAAAC__AAUG_G__AC_A_UUUUAGGAAAAAUAUUUUCAAUAAACUUU ((.((((((.(.(((((((.(......)))))))).)..)).)))).))....((((((((.................))))))))..................... (-17.45 = -18.10 + 0.64)

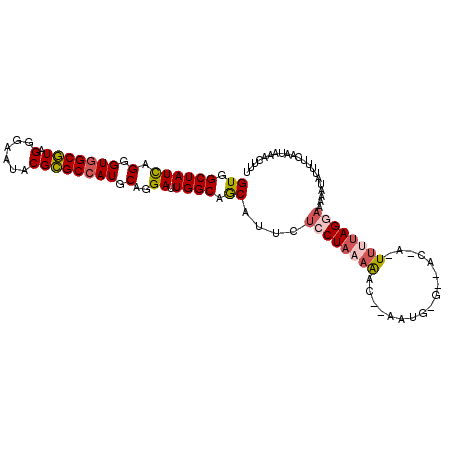

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:07:10 2006