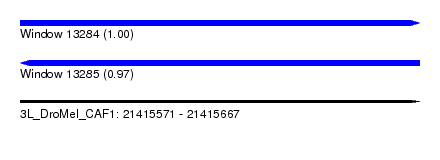
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,415,571 – 21,415,667 |
| Length | 96 |
| Max. P | 0.999740 |

| Location | 21,415,571 – 21,415,667 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 70.12 |
| Mean single sequence MFE | -32.35 |
| Consensus MFE | -20.69 |
| Energy contribution | -19.75 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.64 |
| SVM decision value | 3.98 |
| SVM RNA-class probability | 0.999740 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
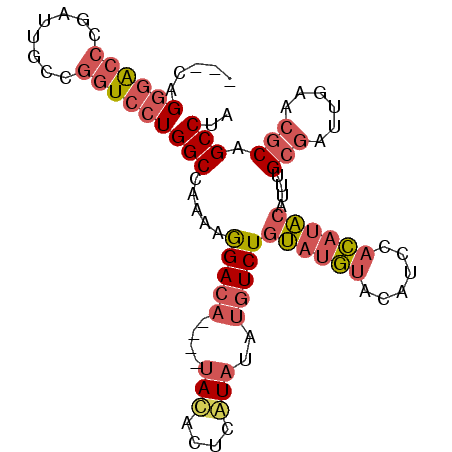
>3L_DroMel_CAF1 21415571 96 + 23771897 UAGGCUGCGUUCAAUCGCAAAUGUAUGUGGAUGUACAUACAGACAUAUAUAUAUAUAUAUCUGUCCUUUUGGCCAGGACCGGCAAUCGGGUCCCUG--- ..(((((((......)))...((((((((....))))))))((((.(((((....))))).)))).....)))).(((((.(....).)))))...--- ( -31.00) >DroSec_CAF1 6998 92 + 1 UAGGCUGCGUUCAAUCGCAAAUGUAUGUGGAUGUACAUACAGACAUAUACGAGUGUA----UGUCCUUUUGGCCAGGACCGGCAACCGGGUCCCUG--- ..(((((((......)))...((((((((....))))))))((((((((....))))----)))).....)))).((((((....)..)))))...--- ( -34.10) >DroSim_CAF1 7215 92 + 1 UAGGCUGCGUUCAAUCGCAAAUGUAUGUGGAUGUACAUACAGACAUAUACGAGUGUA----UGUCCUUUUGGCCAGGACCGGCAACCGGGUCCCUG--- ..(((((((......)))...((((((((....))))))))((((((((....))))----)))).....)))).((((((....)..)))))...--- ( -34.10) >DroAna_CAF1 7721 86 + 1 CAGGCAGUG-------GCA-GUGGCUCUGGCUCUGGCUCUAGAU----AUGACUGUGU-CCUGUCUCUAUCGCCAGGAUUCAGAUUCAGGCUUCUGUAG ...((((.(-------((.-.((..(((((.((((((..((((.----..(((.....-...)))))))..))))))..)))))..)).))).)))).. ( -30.20) >consensus UAGGCUGCGUUCAAUCGCAAAUGUAUGUGGAUGUACAUACAGACAUAUACGAGUGUA____UGUCCUUUUGGCCAGGACCGGCAACCGGGUCCCUG___ ..(((((((......)))...((((((((....))))))))((((..(((....)))....)))).....)))).(((((........)))))...... (-20.69 = -19.75 + -0.94)



| Location | 21,415,571 – 21,415,667 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 70.12 |
| Mean single sequence MFE | -28.35 |
| Consensus MFE | -13.85 |
| Energy contribution | -15.97 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972123 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21415571 96 - 23771897 ---CAGGGACCCGAUUGCCGGUCCUGGCCAAAAGGACAGAUAUAUAUAUAUAUAUGUCUGUAUGUACAUCCACAUACAUUUGCGAUUGAACGCAGCCUA ---..((((((........))))))(((.....(((((.(((((....))))).)))))((((((......))))))...((((......))))))).. ( -29.70) >DroSec_CAF1 6998 92 - 1 ---CAGGGACCCGGUUGCCGGUCCUGGCCAAAAGGACA----UACACUCGUAUAUGUCUGUAUGUACAUCCACAUACAUUUGCGAUUGAACGCAGCCUA ---..((((((.((...))))))))(((.....(((((----((........)))))))((((((......))))))...((((......))))))).. ( -29.70) >DroSim_CAF1 7215 92 - 1 ---CAGGGACCCGGUUGCCGGUCCUGGCCAAAAGGACA----UACACUCGUAUAUGUCUGUAUGUACAUCCACAUACAUUUGCGAUUGAACGCAGCCUA ---..((((((.((...))))))))(((.....(((((----((........)))))))((((((......))))))...((((......))))))).. ( -29.70) >DroAna_CAF1 7721 86 - 1 CUACAGAAGCCUGAAUCUGAAUCCUGGCGAUAGAGACAGG-ACACAGUCAU----AUCUAGAGCCAGAGCCAGAGCCAC-UGC-------CACUGCCUG ...(((..((.((..((((..((.((((..((((.....(-((...)))..----.))))..))))))..))))..)).-.))-------..))).... ( -24.30) >consensus ___CAGGGACCCGAUUGCCGGUCCUGGCCAAAAGGACA____UACACUCAUAUAUGUCUGUAUGUACAUCCACAUACAUUUGCGAUUGAACGCAGCCUA .....((((((........))))))(((.....(((((....(((....)))..)))))((((((......))))))....(((......))).))).. (-13.85 = -15.97 + 2.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:07:02 2006