
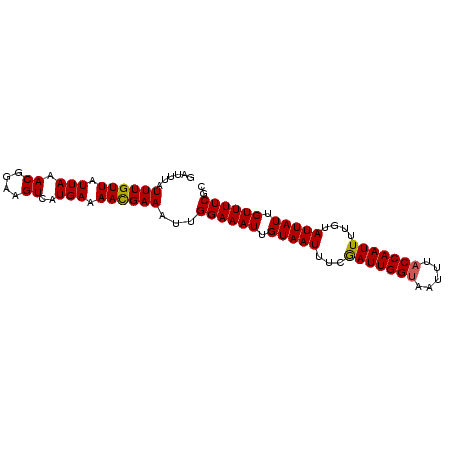
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,395,333 – 21,395,425 |
| Length | 92 |
| Max. P | 0.881689 |

| Location | 21,395,333 – 21,395,425 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 96.20 |
| Mean single sequence MFE | -20.02 |
| Consensus MFE | -17.33 |
| Energy contribution | -17.20 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
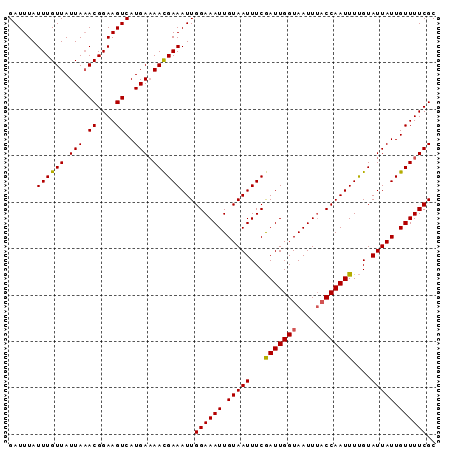
>3L_DroMel_CAF1 21395333 92 + 23771897 GAUUUAUUUGUUAUUAAACGGAAGUCAUGAAAACGAAAUUGGAAAUUGUAAUUUCGAUUGGUAAUUUACCAAUUUUGUAUUAUUGUUUUCGC ......((((((.(((.((....))..))).))))))...((((((.(((((...((((((((...))))))))....))))).)))))).. ( -19.40) >DroSec_CAF1 28043 92 + 1 GAUUUAUUUGUUAUUAAACGGAAGUCAUGAAAACGAAAUUGGAAAUUGUAAUUUCGAUUGGUAAUUUACCAAUUUUGUAUUAUUGUUUUCGC ......((((((.(((.((....))..))).))))))...((((((.(((((...((((((((...))))))))....))))).)))))).. ( -19.40) >DroSim_CAF1 28145 89 + 1 GAUUUAUUUGUUAUUA-ACGGAAGUCAUGA-AACGAAAUUGGAAAUUGUAAUUUCGAUUGGUAAUUU-CCAAUUUUGUAUUAUUGUUUUCGC ................-.((((((..((((-.((((((((((((((((..(.......)..))))))-)))))))))).))))..)))))). ( -23.40) >DroYak_CAF1 28954 91 + 1 GAUUUAUUUGUUAUUAAACGGAAGUCAUGAAAAUGAAAUUGGAAAUUGUAAUUUCAAUUGGUAAUU-ACCAAUUUCGUAUUAUUGUUUUCGC ......((((((....))))))..((((....))))....((((((.(((((...(((((((....-)))))))....))))).)))))).. ( -17.90) >consensus GAUUUAUUUGUUAUUAAACGGAAGUCAUGAAAACGAAAUUGGAAAUUGUAAUUUCGAUUGGUAAUUUACCAAUUUUGUAUUAUUGUUUUCGC ......((((((.(((.((....))..))).))))))...((((((.(((((...(((((((.....)))))))....))))).)))))).. (-17.33 = -17.20 + -0.13)

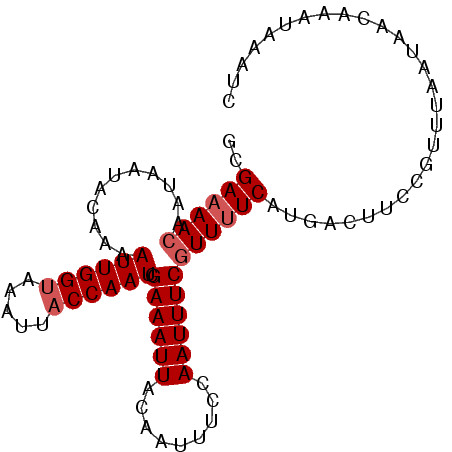
| Location | 21,395,333 – 21,395,425 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 96.20 |
| Mean single sequence MFE | -12.32 |
| Consensus MFE | -9.95 |
| Energy contribution | -10.70 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.606107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21395333 92 - 23771897 GCGAAAACAAUAAUACAAAAUUGGUAAAUUACCAAUCGAAAUUACAAUUUCCAAUUUCGUUUUCAUGACUUCCGUUUAAUAACAAAUAAAUC ..((((((...........(((((((...))))))).((((((.........)))))))))))).........(((....)))......... ( -13.70) >DroSec_CAF1 28043 92 - 1 GCGAAAACAAUAAUACAAAAUUGGUAAAUUACCAAUCGAAAUUACAAUUUCCAAUUUCGUUUUCAUGACUUCCGUUUAAUAACAAAUAAAUC ..((((((...........(((((((...))))))).((((((.........)))))))))))).........(((....)))......... ( -13.70) >DroSim_CAF1 28145 89 - 1 GCGAAAACAAUAAUACAAAAUUGG-AAAUUACCAAUCGAAAUUACAAUUUCCAAUUUCGUU-UCAUGACUUCCGU-UAAUAACAAAUAAAUC .................(((((((-(((((...............)))))))))))).(((-...((((....))-))..)))......... ( -11.06) >DroYak_CAF1 28954 91 - 1 GCGAAAACAAUAAUACGAAAUUGGU-AAUUACCAAUUGAAAUUACAAUUUCCAAUUUCAUUUUCAUGACUUCCGUUUAAUAACAAAUAAAUC ..(((((...........(((((((-....)))))))((((((.........)))))).))))).........(((....)))......... ( -10.80) >consensus GCGAAAACAAUAAUACAAAAUUGGUAAAUUACCAAUCGAAAUUACAAUUUCCAAUUUCGUUUUCAUGACUUCCGUUUAAUAACAAAUAAAUC ..((((((...........((((((.....)))))).((((((.........))))))))))))............................ ( -9.95 = -10.70 + 0.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:06:49 2006