| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,385,557 – 21,385,654 |
| Length | 97 |
| Max. P | 0.947299 |

| Location | 21,385,557 – 21,385,654 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 77.89 |
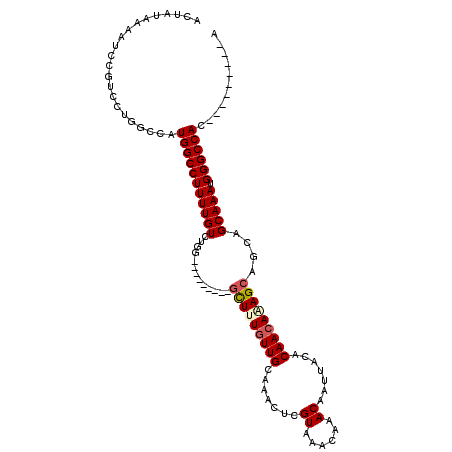
| Mean single sequence MFE | -26.60 |
| Consensus MFE | -16.84 |
| Energy contribution | -17.17 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
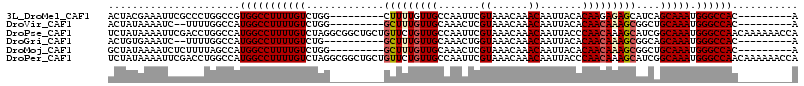
>3L_DroMel_CAF1 21385557 97 - 23771897 ACUACGAAAUUCGCCCUGGCCGUGGCCUUUUGUCUGG---------CUUUUGUUGCCAAUUCGUAAACAAACAAUUACACAAGAGAGCAUCAGCAAAUGGGCCAC---------A ............((....)).(((((((((((.((((---------(((((..((..((((.((......)))))).))...))))))..))))))).)))))))---------. ( -26.10) >DroVir_CAF1 5027 95 - 1 ACUAUAAAAUC--UUUUGGCCAUGGCCUUUUGUCUGG---------GCUUUGUUGCAAACUCGUAAACAAACAAUUACACAACAAAGCGGCUGCAAAUGGGCCAC---------A ...........--.........(((((((((((....---------(((((((((.......((......)).......)))))))))....))))).)))))).---------. ( -23.74) >DroPse_CAF1 4912 115 - 1 UCUAUAAAAUUCGACCUGGCCAUGGCCUUUUGUCUAGGCGGCUGCUGUUCUGUUGCCAAUUCGUAAACAAACAAUUACCCAACAAAGCAUCGGCAAAUGGGCCAACAAAAAACCA ................(((((...((((.......)))).((((.((((.(((((..((((.((......))))))...))))).)))).)))).....)))))........... ( -28.80) >DroGri_CAF1 5569 94 - 1 ACUGUGAAAUC--UUUUGGCCAUGGCCUUUUGUCUG----------GCUUUGUUGCAAACUGGUAAACAAACAAUUACACAACAAAGCGGCAGCAAAUGGGCCAC---------A ...........--.........((((((((((.(((----------(((((((((.......((......)).......)))))))))..))))))).)))))).---------. ( -26.34) >DroMoj_CAF1 5029 97 - 1 GCUAUAAAAUCUCUUUUAGCCAUGGCCUUUUGUCUGG---------GCUUUGUUGCAAACUCGUAAACAAACAAUUACACAACAAAGCGGCUGCAAAUGGGCCAC---------A ((((.((......)).))))..(((((((((((....---------(((((((((.......((......)).......)))))))))....))))).)))))).---------. ( -25.84) >DroPer_CAF1 4925 115 - 1 UCUAUAAAAUUCGACCUGGCCAUGGCCUUUUGUCUAGGCGGCUGCUGUUCUGUUGCCAAUUCGUAAACAAACAAUUACCCAACAAAGCAUCGGCAAAUGGGCCAACAAAAAACCA ................(((((...((((.......)))).((((.((((.(((((..((((.((......))))))...))))).)))).)))).....)))))........... ( -28.80) >consensus ACUAUAAAAUCCGUCCUGGCCAUGGCCUUUUGUCUGG_________GCUUUGUUGCAAACUCGUAAACAAACAAUUACACAACAAAGCAGCAGCAAAUGGGCCAC_________A ......................(((((((((((.............(((((((((.......((......)).......)))))))))....))))).))))))........... (-16.84 = -17.17 + 0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:06:47 2006