
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,384,431 – 21,384,523 |
| Length | 92 |
| Max. P | 0.808834 |

| Location | 21,384,431 – 21,384,523 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 79.44 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -18.70 |
| Energy contribution | -18.73 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
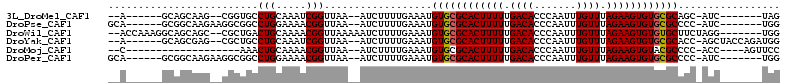
>3L_DroMel_CAF1 21384431 92 - 23771897 --A------GCAGCAAG--CGGUGCCUGCAAAUCGGUUAA--AUCUUUUGAAAUGUGCGCACUUUUUGACACCCAAUUUGUUUAGAAGUGUGCGCAGC-AUC-------UAG --.------((.....)--)(((((...((((........--....))))...(((((((((((((.((((.......)))).)))))))))))))))-)))-------... ( -28.40) >DroPse_CAF1 3898 96 - 1 GCA------GCGGCAAGAAGGCGGCCUGGAAAACGGUUAA--AUCUUUUGAAAUGUGCGCACUUUUUGACACCCAAUUUGUUUAGAAGUGUGCGCCCC-AUC-------UGG ((.------...)).(((.((.((((((.....)))....--..............((((((((((.((((.......)))).)))))))))))))))-.))-------).. ( -28.40) >DroWil_CAF1 6652 101 - 1 --ACCAAAGGCAGCAGC--CGCUGACUGCAAAACGGUUAAAAAUCUUUUGAAAUGUGCGCACUUUUUGACACCCAAUUUGUUUAGAAGUGUGUGCUUCUAGG-------UGG --(((....(((((((.--..))).)))).....))).....((((...(((..(..(((((((((.((((.......)))).)))))))))..)))).)))-------).. ( -28.70) >DroYak_CAF1 17937 99 - 1 --A------GCAGCGAG--CGCUGCCUGCAAAUCGGUUAA--AUCUUUUGAAAUGUGCGCACUUUUUGACACCCAAUUUGUUUAGAAGUGUGCGCACC-AGCUACCAGAUGG --.------(((((...--.)))))......((((((...--.((....))..(((((((((((((.((((.......)))).)))))))))))))..-....))).))).. ( -32.10) >DroMoj_CAF1 3994 84 - 1 --C-------------------AAACUGCAAAACGGUUAA--AUCUUUUGAAAUGUGCGCACUUUUUGACACCCAAUUUGUUUAGAAGUGUACGCCCC-ACC----AGUUCC --.-------------------.((((((((((.(((...--))))))))....(((..(((((((.((((.......)))).)))))))..)))...-..)----)))).. ( -18.40) >DroPer_CAF1 3907 96 - 1 GCA------GCGGCAAGAAGGCGGCCUGGAAAACGGUUAA--AUCUUUUGAAAUGUGCGCACUUUUUGACACCCAAUUUGUUUAGAAGUGUGCGCCCC-AUC-------UGG ((.------...)).(((.((.((((((.....)))....--..............((((((((((.((((.......)))).)))))))))))))))-.))-------).. ( -28.40) >consensus __A______GCAGCAAG__CGCUGCCUGCAAAACGGUUAA__AUCUUUUGAAAUGUGCGCACUUUUUGACACCCAAUUUGUUUAGAAGUGUGCGCCCC_AUC_______UGG .........................(((.....)))..................((((((((((((.((((.......)))).))))))))))))................. (-18.70 = -18.73 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:06:46 2006