
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,369,296 – 21,369,474 |
| Length | 178 |
| Max. P | 0.999999 |

| Location | 21,369,296 – 21,369,409 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 74.11 |
| Mean single sequence MFE | -51.68 |
| Consensus MFE | -27.11 |
| Energy contribution | -26.92 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21369296 113 - 23771897 CAGCUGCCAUAGCCGCUGCUUGGGCGGCACUAUGCGCCUCCGCCGAGAUCCCGCUUAAGUGCAUCUGGAAGGGCAAGGGGAGUUCCGUGGGUGGCGGCGGCA-CAGAGGGUGGU--- ..((..((...((((((.....)))))).((.((.(((.((((((....(((((.....(((.((.....)))))...((....))))))))))))).))).-)).))))..))--- ( -50.20) >DroPse_CAF1 19525 113 - 1 CAGCCGCCAUUGCUGCAGCCUGAGCGGCACUAUGCGCCUCGGCCGAAAUCCCAGUCAGAUGCAUUUGGAAGGGUAAGGCAAGUUCCGCUGGAGCUGGAGGCG-CUGAUGGUGGU--- ..((((((((((((((.......))))).....(((((((.(((.....(((..(((((....)))))..)))...))).((((((...)))))).))))))-).)))))))))--- ( -54.60) >DroGri_CAF1 51358 105 - 1 CAGCUGCCAUUGCGGCCGCCUGUGCGGCACUGUGUGCCUCCGCUGAAAUUCCGCUCAAAUGCAUAUUAAAGGGUAAGGCGAUUUCUGUCGGCGCGGGCGGCUGUU------------ ...........(((((((((((((((((((...)))))..(((.((((((((((......)).((((....)))).)).)))))).).)))))))))))))))).------------ ( -45.20) >DroMoj_CAF1 34757 114 - 1 CAGCUGCCAUUGCGGCCGCUUGAGCGGCACUGUGAGCCUCCGCUGAAAUUCCGCUCAAAUGCAUG---AAGGGAAGGGAUACAUCUGUGGGCGCGGGCGGCUGUUGAUUGUGUUGUU ((((((((...(((.((((((((((((.....(.(((....))).)....)))))))).......---...(((.(.....).))))))).))).)))))))).............. ( -45.00) >DroAna_CAF1 30637 113 - 1 CGGCUGCCAUGGCCGCUGCUUGGGCGGCACUGUGCGCCUCCGCCGAGAUCCCGGUUAGGUGCAUUUGGAAGGGCAGUGGCAGCUCCGUGGGCGGCGGCGGUG-CUGGCGGAGGU--- ..((((((((.((((((((....))))).((((((((((..((((......)))).))))))))..))...))).))))))))(((((.((((.(...).))-)).)))))...--- ( -60.50) >DroPer_CAF1 21442 113 - 1 CAGCCGCCAUUGCUGCAGCCUGAGCGGCACUAUGCGCCUCGGCCGAAAUCCCAGUCAGAUGCAUUUGGAAGGGUAAGGCAAGUUCCGCUGGAGCUGGAGGCG-CUGAUGGUGGU--- ..((((((((((((((.......))))).....(((((((.(((.....(((..(((((....)))))..)))...))).((((((...)))))).))))))-).)))))))))--- ( -54.60) >consensus CAGCUGCCAUUGCCGCAGCCUGAGCGGCACUAUGCGCCUCCGCCGAAAUCCCGCUCAAAUGCAUUUGGAAGGGUAAGGCAAGUUCCGUGGGCGCCGGCGGCG_CUGAUGGUGGU___ ..((((((......(((((....))))).......((..((((.(((..(((..................))).........))).))))..)).))))))................ (-27.11 = -26.92 + -0.19)

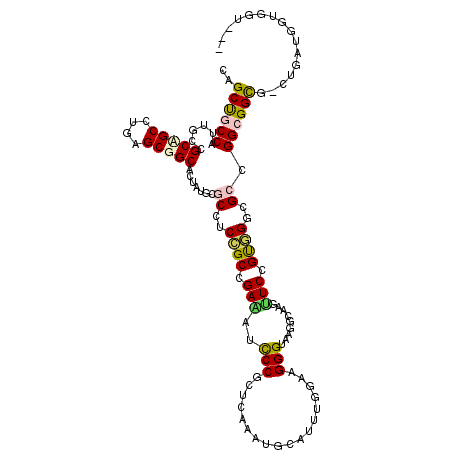

| Location | 21,369,329 – 21,369,449 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.22 |
| Mean single sequence MFE | -51.73 |
| Consensus MFE | -30.29 |
| Energy contribution | -31.07 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.727415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21369329 120 + 23771897 CCCCUUGCCCUUCCAGAUGCACUUAAGCGGGAUCUCGGCGGAGGCGCAUAGUGCCGCCCAAGCAGCGGCUAUGGCAGCUGCCCAAGCUGCAGCAGCCCAAGCUGCUGCGGCGGAACAACA ......(((((.((((((.(.(......)).)))).)).)).))).....((.(((((...(((((((((..(((.(((((.......))))).)))..)))))))))))))).)).... ( -53.60) >DroPse_CAF1 19558 120 + 1 UGCCUUACCCUUCCAAAUGCAUCUGACUGGGAUUUCGGCCGAGGCGCAUAGUGCCGCUCAGGCUGCAGCAAUGGCGGCUGCCCAGGCCGCGGCGGCUCAGGCGGCAGCGGCAGACCAGCA .................(((......(((((....(((((..(((((...)))))(((...((....))...)))))))))))))(((((.((.((....)).)).)))))......))) ( -49.60) >DroWil_CAF1 18874 120 + 1 UACGUUACCCUUCAAUAUGCAUCUAGCUGGUAUUUCACCAGGUGCCCAUAGUGCCGCCCAAGCGGCAGCCAUGGCCGCUGCCCAAGCAGCCGCCGCUCAGGCUGCUGCAGCUGAACAGCA .................(((...((((((((.....))).((..(.....)..))......(((((((((.((((.(((((....))))).))))....))))))))))))))....))) ( -46.60) >DroMoj_CAF1 34794 117 + 1 AUCCCUUCCCUU---CAUGCAUUUGAGCGGAAUUUCAGCGGAGGCUCACAGUGCCGCUCAAGCGGCCGCAAUGGCAGCUGCGCAGGCCGCGGCAGCGCAAGCCGCCGCUGCGGAUCAACA ((((........---..(((..((((((((.(((..(((....)))...))).))))))))(((((.((....)).)))))))).((.(((((.((....)).))))).))))))..... ( -53.00) >DroAna_CAF1 30670 118 + 1 GCCACUGCCCUUCCAAAUGCACCUAACCGGGAUCUCGGCGGAGGCGCACAGUGCCGCCCAAGCAGCGGCCAUGGCAGCCGCCCAAGCUGCAGCGGCACAGGCAGCGGCGGCCGAACAG-- (((.(((((........(((.(((..((((....))))...))).)))..(((((((...(((.(((((.......)))))....)))...))))))).))))).)))..........-- ( -55.40) >DroPer_CAF1 21475 120 + 1 UGCCUUACCCUUCCAAAUGCAUCUGACUGGGAUUUCGGCCGAGGCGCAUAGUGCCGCUCAGGCUGCAGCAAUGGCGGCUGCCCAGGCCGCGGCGGCUCAGGCGGCAGCGGCCGACCAGCA .((....(((...((........))...)))...(((((((..((((...(.((((((..(((((((((.......)))))...))))..)))))).)..)).))..)))))))...)). ( -52.20) >consensus UCCCUUACCCUUCCAAAUGCAUCUAACCGGGAUUUCGGCGGAGGCGCAUAGUGCCGCCCAAGCAGCAGCAAUGGCAGCUGCCCAAGCCGCGGCGGCUCAGGCGGCAGCGGCCGAACAGCA ............................(((....((((...(((((...)))))(((...((....))...))).)))))))..(((((.((.((....)).)).)))))......... (-30.29 = -31.07 + 0.78)



| Location | 21,369,369 – 21,369,474 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.96 |
| Mean single sequence MFE | -50.37 |
| Consensus MFE | -31.54 |
| Energy contribution | -31.77 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.63 |
| SVM decision value | 6.90 |
| SVM RNA-class probability | 0.999999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21369369 105 + 23771897 GAGGCGCAUAGUGCCGCCCAAGCAGCGGCUAUGGCAGCUGCCCAAGCUGCAGCAGCCCAAGCUGCUGCGGCGGAACAACAGCAACCGCCGC---------------CACCAACUUCACAU ..((((....((.(((((...(((((((((..(((.(((((.......))))).)))..)))))))))))))).))....((....)))))---------------)............. ( -48.40) >DroGri_CAF1 51423 120 + 1 GAGGCACACAGUGCCGCACAGGCGGCCGCAAUGGCAGCUGCGCAGGCCGCGGCAGCGCAAGCCGCCGCCGCGGAUCAACAGCAGCCGGCGGCAGCGCCUCCAUCGGCAUCAUCAUCGCAU ..(((((...)))))((....((.(((((...(((.((((......(((((((.(((.....))).))))))).....)))).))).))))).))(((......))).........)).. ( -60.20) >DroWil_CAF1 18914 99 + 1 GGUGCCCAUAGUGCCGCCCAAGCGGCAGCCAUGGCCGCUGCCCAAGCAGCCGCCGCUCAGGCUGCUGCAGCUGAACAGCA------ACCGC---------------CUCCCUCAUCACAU ((..(.....)..))((....(((((((((.((((.(((((....))))).))))....))))))))).(((....))).------...))---------------.............. ( -42.50) >consensus GAGGCACAUAGUGCCGCCCAAGCGGCAGCAAUGGCAGCUGCCCAAGCAGCAGCAGCCCAAGCUGCUGCAGCGGAACAACAGCA_CCGCCGC_______________CACCAUCAUCACAU ..........((.((((....(((((((((.((((.(((((.......))))).)).))))))))))).))))....))......................................... (-31.54 = -31.77 + 0.23)


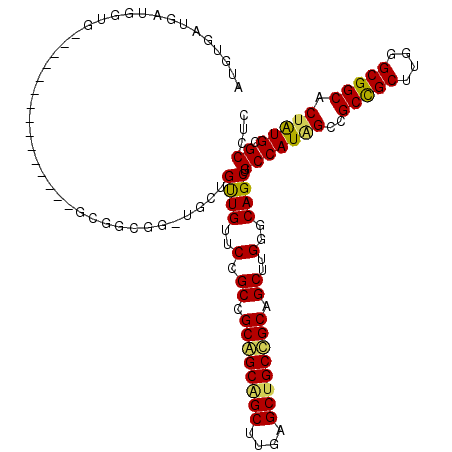
| Location | 21,369,369 – 21,369,474 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 66.96 |
| Mean single sequence MFE | -59.80 |
| Consensus MFE | -37.50 |
| Energy contribution | -37.40 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.22 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.63 |
| SVM decision value | 6.88 |
| SVM RNA-class probability | 0.999999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21369369 105 - 23771897 AUGUGAAGUUGGUG---------------GCGGCGGUUGCUGUUGUUCCGCCGCAGCAGCUUGGGCUGCUGCAGCUUGGGCAGCUGCCAUAGCCGCUGCUUGGGCGGCACUAUGCGCCUC ..(((....(((((---------------.((((((..((....)).)))))((((((((....)))))))).(((..((((((.((....)).))))))..)))).)))))..)))... ( -59.20) >DroGri_CAF1 51423 120 - 1 AUGCGAUGAUGAUGCCGAUGGAGGCGCUGCCGCCGGCUGCUGUUGAUCCGCGGCGGCGGCUUGCGCUGCCGCGGCCUGCGCAGCUGCCAUUGCGGCCGCCUGUGCGGCACUGUGUGCCUC ..(((...((..(((((.((.(((((..(((((.(((.(((((....(((((((((((.....))))))))))).....))))).)))...)))))))))).)))))))..)).)))... ( -67.30) >DroWil_CAF1 18914 99 - 1 AUGUGAUGAGGGAG---------------GCGGU------UGCUGUUCAGCUGCAGCAGCCUGAGCGGCGGCUGCUUGGGCAGCGGCCAUGGCUGCCGCUUGGGCGGCACUAUGGGCACC .........((.((---------------(((((------(((((((((((((...))).))))))))))))))))).......(.((((((.((((((....)))))))))))).).)) ( -52.90) >consensus AUGUGAUGAUGGUG_______________GCGGCGG_UGCUGUUGUUCCGCCGCAGCAGCUUGAGCUGCCGCAGCUUGGGCAGCUGCCAUAGCCGCCGCUUGGGCGGCACUAUGCGCCUC .........................................((((..(.((.((((((((....)))))))).))..)..)))).(((((((..(((((....))))).))))).))... (-37.50 = -37.40 + -0.10)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:06:44 2006