
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,354,929 – 21,355,033 |
| Length | 104 |
| Max. P | 0.680762 |

| Location | 21,354,929 – 21,355,033 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.94 |
| Mean single sequence MFE | -31.12 |
| Consensus MFE | -17.80 |
| Energy contribution | -17.42 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.680762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
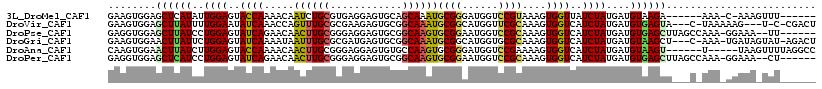
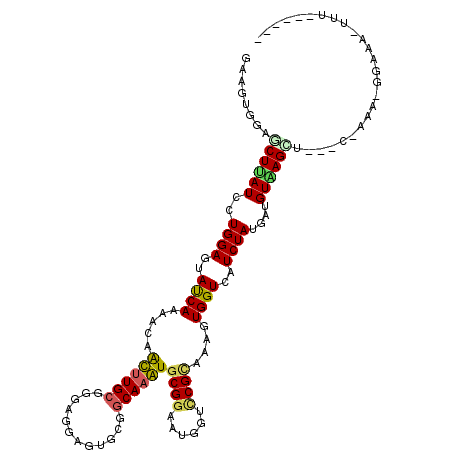
>3L_DroMel_CAF1 21354929 104 - 23771897 GAAGUGGAGCUCAUAUUGGAGUACCAAAACAAUCUGCGUGAGGAGUGCAGCAAAUGCGGGAUGGUCCGUAAAGUGGUUAUCUAUGAUGUAAGA------AAA-C-AAAGUUU------ ....(((.((((......)))).)))((((...((.(....).))((((((....)).((((((.(((.....)))))))))....))))...------...-.-...))))------ ( -21.80) >DroVir_CAF1 18567 109 - 1 GAAGUGGAGCUUAUUUUGGAAUAUCAAACCAGUUUGCGCGAAGAGUGCGGCAAAUGCGGCAUGGUUCGCAAAGUGGUCAUCUAUGAUGUGAGUA---C-UAAAAAG---U-C-CGACU ..(((.(..(((..(((((....(((.((((.((((((.((...((((.((....)).))))..)))))))).))))((((...)))))))...---)-)))))))---.-.-).))) ( -32.90) >DroPse_CAF1 1222 109 - 1 GAGGUGGAGCUUAUCCUGGAGUAUCAGAACAACUUGCGGGAGGAGUGCGGCAAGUGCGGAAUGGUCCGCAAAGUGGUCAUCUAUGAUGUGAGCUUAGCCAAA-GGAAA--UU------ ..(((.((((((((((((((((.........)))).)))))(((..((.((...((((((....))))))..)).))..)))......))))))).)))...-.....--..------ ( -35.30) >DroGri_CAF1 34484 112 - 1 GAAGUGGAACUUAUUCUGGAGUAUCAAAAUAAUUUGCGCGAUGAGUGCGGCAAAUGCGGCAUGGUGCGCAAAGUGGUCAUCUAUGAUGUAAGCU---C-AAA-UGAUAGUAU-AGACU ..............((((.(.(((((......((((((((....((((.((....)).))))..))))))))((...((((...))))...)).---.-...-))))).).)-))).. ( -31.00) >DroAna_CAF1 16437 107 - 1 CAAGUGGAACUUAUCUUGGAGUACCAAAACAACUUGCGGGAGGAGUGUGCCAAGUGCGGGAUGGUCCGAAAAGUGGUCAUCUAUGAUGUAAGU------U-----UAAGUUUUAGGCC ....(((((((((..((((....))))...((((((((...(((.((.((((....((((....)))).....)))))))))....)))))))------)-----))))))))).... ( -26.50) >DroPer_CAF1 1225 109 - 1 GAGGUGGAGCUCAUCCUGGAGUAUCAGAACAACUUGCGGGAGGAGUGCGGCAAGUGCGGAAUGGUCCGCAAAGUGGUCAUCUAUGAUGUGAGCUUAGCCAAA-GGAAA--CU------ ..(((.((((((((((((((((.........)))).)))))(((..((.((...((((((....))))))..)).))..)))......))))))).)))..(-(....--))------ ( -39.20) >consensus GAAGUGGAGCUUAUCCUGGAGUAUCAAAACAACUUGCGGGAGGAGUGCGGCAAAUGCGGAAUGGUCCGCAAAGUGGUCAUCUAUGAUGUAAGCU___C_AAA_GGAAA_UUU______ ........((((((..((((..((((.....((((((............))))))((((......))))....))))..))))....))))))......................... (-17.80 = -17.42 + -0.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:06:38 2006