| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,342,752 – 21,342,859 |
| Length | 107 |
| Max. P | 0.994666 |

| Location | 21,342,752 – 21,342,859 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 75.76 |
| Mean single sequence MFE | -38.45 |
| Consensus MFE | -24.47 |
| Energy contribution | -24.50 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21342752 107 - 23771897 UCAAUGAAAUCCAGACAGUUUCU--CGCCUAGAUGCAUUUGGCCUGCGUGGGCCUAACGGUCCAGCAAAAGUGCAUC-UGCGAU--GAGGAACGAGGCGAUGGAUGAAGGAA ((......(((((..(.((((((--(((.(((((((((((....(((.((((((....))))))))).)))))))))-)).).)--)))))))...)...)))))....)). ( -44.40) >DroSec_CAF1 45644 106 - 1 UCUAUGAAAUCCAGACAGU-UCU--CGCCAAGAUGCAUUUGGCCUGCGUGGGCCUAACGGUCCAGCAAAAGUGCAUC-UGCGAU--GAGGAACGAGGCGAUGUAUGGAGGAA .........((((.(((..-..(--((((.((((((((((....(((.((((((....))))))))).)))))))))-).((..--......)).)))))))).)))).... ( -41.70) >DroSim_CAF1 46821 106 - 1 UCUAUGAAAUCCAGACAGU-UCU--CGCCAAGAUGCAUUUGGCCUGCGUGGGCCUAACGGUCCAGCAAAAGUGCAUC-UGCGAU--GAGGAACGAGGCGAUGGAUGGAGGAA .........((((..((..-..(--((((.((((((((((....(((.((((((....))))))))).)))))))))-).((..--......)).)))))))..)))).... ( -42.30) >DroEre_CAF1 47759 85 - 1 UCUAUGAAAUCCCGACAGU--CU--CGCCAAGAUGCAUUUGGCCUGCGUGGGCCUAACGGUCCGGCAAAAAUGCAUC-UGCGCU--GAG--------------------GAA .........(((...(((.--..--(((..((((((((((....(((.((((((....))))))))).)))))))))-))))))--).)--------------------)). ( -33.20) >DroYak_CAF1 73224 105 - 1 UCUAUGAAAUCCAGACAGU--CU--CGCCAAGAUGCAUUUGGCCUGCGUGGGGCUAACGGUUCAGCAAAAAUGCAUC-UGCGAU--GAGGAACGAGGCGAUGGAUGGAGGAA .........((((..((..--.(--((((.((((((((((....(((.((((.(....).))))))).)))))))))-).((..--......)).)))))))..)))).... ( -35.10) >DroPer_CAF1 65254 91 - 1 ACUCGGAG-------GAGU-UCUAAGGCGAAAAUGCGAUUCGCUGGCGUGGGCUUAACGGUCCAGA--GAUUGCCCCGAGCGACUGGAGUCUCGAGUCG-----------AG .(((((.(-------(...-(((..((((((.......))))))....((((((....)))))).)--))...)))))))(((((.(.....).)))))-----------.. ( -34.00) >consensus UCUAUGAAAUCCAGACAGU_UCU__CGCCAAGAUGCAUUUGGCCUGCGUGGGCCUAACGGUCCAGCAAAAGUGCAUC_UGCGAU__GAGGAACGAGGCGAUGGAUGGAGGAA .........................(((...(((((((((....(((.((((((....))))))))).)))))))))..))).............................. (-24.47 = -24.50 + 0.03)


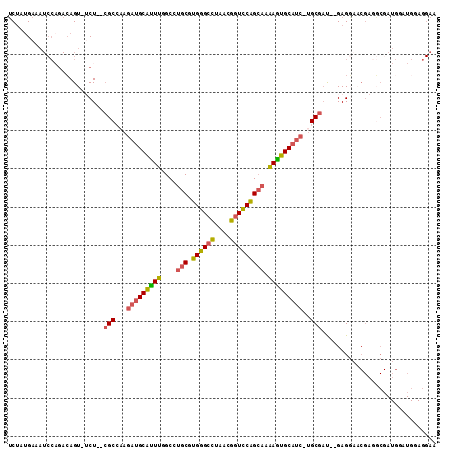
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:06:31 2006