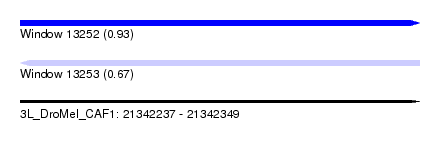
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,342,237 – 21,342,349 |
| Length | 112 |
| Max. P | 0.925861 |

| Location | 21,342,237 – 21,342,349 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 87.22 |
| Mean single sequence MFE | -24.66 |
| Consensus MFE | -19.28 |
| Energy contribution | -19.96 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
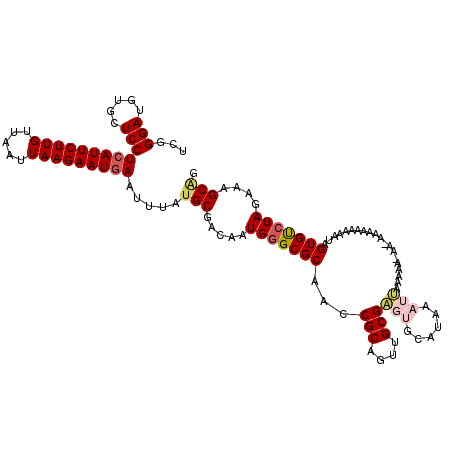
>3L_DroMel_CAF1 21342237 112 + 23771897 CGGCUUUUUAUGCACUAUUUUUUUUUUUUUUUUUUGAUUUA-----CCGCAACUGCGGUUGCGCCCAUUGUCGCAUAAAUUCAUUCUUAAUUAACAAGAAUGAGGAGCACAUCCCGA ..(((((((((((...........................(-----((((....))))).(((.....))).)))))))(((((((((.......)))))))))))))......... ( -24.40) >DroSec_CAF1 45129 116 + 1 CUGCUUUCUAGACACUAUUUUUUAUU-UUCUUUUUAAUUUAUGCACUCGCAACUGCGGUUGCGCCCAUUGUCGCAUAAAUUCAUUCUUAAUUAACAAGAAUGAGGAGCACAUCCCGA .((((((...................-.........((((((((((.((((((....))))))......)).))))))))((((((((.......))))))))))))))........ ( -26.00) >DroSim_CAF1 46306 116 + 1 CUGCUUUCUAGACACUAUUUUUUCUU-UUCUUUUUAAUUUAUGCCAUCGCAACUGCGGUUGCGCCCAUUGUCGCAUAAAUUUAUUCUUAAUUAACAAGAAUGAGGAGCACAUCCCGA .((((((...................-.........((((((((.((((((((....))))))......)).))))))))((((((((.......))))))))))))))........ ( -22.70) >DroEre_CAF1 47253 110 + 1 CCGCUUUCUAGACACUACUUUAUU-------UUUUAAUUUAUGCACUCGCAUUGGCGGUUGCGCCCACUGUCGCAUAAAUUCAUUCUUAAUUAACAAGAAUGAGGAGCACAUCCCGA ..(((((.................-------.....((((((((....(((..((((....))))...))).))))))))((((((((.......)))))))))))))......... ( -24.60) >DroYak_CAF1 72726 109 + 1 CCGCUUUCUAGACACUAUUUCAUG--------UUUAAUUUAUGCACACGCAUCUGCGGUUGCGCCCAUUGUCGCAUAAAUUCAUUCUUAAUUAACAAGAAUGAGGAGCACAUCCCGA ..(((((.((((((........))--------))))(((((((((((((((........)))).....))).))))))))((((((((.......)))))))))))))......... ( -25.60) >consensus CCGCUUUCUAGACACUAUUUUUUUUU_UU_UUUUUAAUUUAUGCACUCGCAACUGCGGUUGCGCCCAUUGUCGCAUAAAUUCAUUCUUAAUUAACAAGAAUGAGGAGCACAUCCCGA ..(((((.............................((((((((...((((((....)))))).........))))))))((((((((.......)))))))))))))......... (-19.28 = -19.96 + 0.68)


| Location | 21,342,237 – 21,342,349 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 87.22 |
| Mean single sequence MFE | -27.54 |
| Consensus MFE | -19.82 |
| Energy contribution | -20.86 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21342237 112 - 23771897 UCGGGAUGUGCUCCUCAUUCUUGUUAAUUAAGAAUGAAUUUAUGCGACAAUGGGCGCAACCGCAGUUGCGG-----UAAAUCAAAAAAAAAAAAAAAAAAUAGUGCAUAAAAAGCCG .(((.(((..((..(((((((((.....))))))))).....((((........))))(((((....))))-----)........................))..)))......))) ( -27.50) >DroSec_CAF1 45129 116 - 1 UCGGGAUGUGCUCCUCAUUCUUGUUAAUUAAGAAUGAAUUUAUGCGACAAUGGGCGCAACCGCAGUUGCGAGUGCAUAAAUUAAAAAGAA-AAUAAAAAAUAGUGUCUAGAAAGCAG .....(((..(((.(((((((((.....)))))))))......(((((..(((......)))..))))))))..))).............-.......................... ( -28.40) >DroSim_CAF1 46306 116 - 1 UCGGGAUGUGCUCCUCAUUCUUGUUAAUUAAGAAUAAAUUUAUGCGACAAUGGGCGCAACCGCAGUUGCGAUGGCAUAAAUUAAAAAGAA-AAGAAAAAAUAGUGUCUAGAAAGCAG ........((((..(((((((((.....))))))).(((((((((.(.......((((((....)))))).).)))))))))........-..................)).)))). ( -23.70) >DroEre_CAF1 47253 110 - 1 UCGGGAUGUGCUCCUCAUUCUUGUUAAUUAAGAAUGAAUUUAUGCGACAGUGGGCGCAACCGCCAAUGCGAGUGCAUAAAUUAAAA-------AAUAAAGUAGUGUCUAGAAAGCGG ((.(((((((((...((((((((.....))))))))((((((((((...((.((((....))))...))...))))))))))....-------.....)))).))))).))...... ( -30.50) >DroYak_CAF1 72726 109 - 1 UCGGGAUGUGCUCCUCAUUCUUGUUAAUUAAGAAUGAAUUUAUGCGACAAUGGGCGCAACCGCAGAUGCGUGUGCAUAAAUUAAA--------CAUGAAAUAGUGUCUAGAAAGCGG ......(((((((.(((((((((.....)))))))))....((......))))))))).((((..((((....))))...(((.(--------(((......)))).)))...)))) ( -27.60) >consensus UCGGGAUGUGCUCCUCAUUCUUGUUAAUUAAGAAUGAAUUUAUGCGACAAUGGGCGCAACCGCAGUUGCGAGUGCAUAAAUUAAAAA_AA_AAAAAAAAAUAGUGUCUAGAAAGCAG ...(((.....)))(((((((((.....))))))))).....(((.....(((((((...(((....)))(((......)))....................)))))))....))). (-19.82 = -20.86 + 1.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:06:30 2006