| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,329,224 – 21,329,314 |
| Length | 90 |
| Max. P | 0.999147 |

| Location | 21,329,224 – 21,329,314 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 66.90 |
| Mean single sequence MFE | -19.67 |
| Consensus MFE | -10.00 |
| Energy contribution | -10.67 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.51 |
| SVM decision value | 3.40 |
| SVM RNA-class probability | 0.999147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
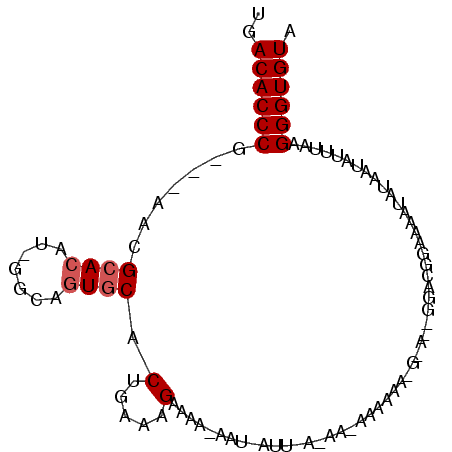
>3L_DroMel_CAF1 21329224 90 + 23771897 UGACACCCG---AACGCACAU-GGCAGUGCACUGAAAGAAAAGAAU-GUUAAUAACAAAAAAUGUA--UGAGGGAAAAUAUAAUAUUUAAGGGUGUA ..((((((.---(((((((..-....)))).((........))...-)))............((((--(........)))))........)))))). ( -18.30) >DroSim_CAF1 33216 72 + 1 UGACACCCG---AACGCACAU-GGCAGUGCACUGAAAGAAAAUAAU-C--------------------GGUCGGAUCAUAUAAUAUUUAAGGGUGUA ..((((((.---...((((..-....)))).((((..((......)-)--------------------..))))................)))))). ( -21.80) >DroYak_CAF1 58704 92 + 1 UGACACCCUGUCAGUGCACAGGGGCACUCCACUGAAAGAAA-----AAUUCAAAAAAAAAAAGGAACUGGGCGGAAAAUAUUAUAUUUAAGGGUGUA ..(((((((((((((((......)))))....((((.....-----..)))).................)))...(((((...))))).))))))). ( -18.90) >consensus UGACACCCG___AACGCACAU_GGCAGUGCACUGAAAGAAAA_AAU_AUU_A_AA_AAAAAA_G_A__GGACGGAAAAUAUAAUAUUUAAGGGUGUA ..((((((.......((((.......)))).(.....)....................................................)))))). (-10.00 = -10.67 + 0.67)

| Location | 21,329,224 – 21,329,314 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 66.90 |
| Mean single sequence MFE | -17.55 |
| Consensus MFE | -9.17 |
| Energy contribution | -10.07 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.52 |
| SVM decision value | 3.29 |
| SVM RNA-class probability | 0.998935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
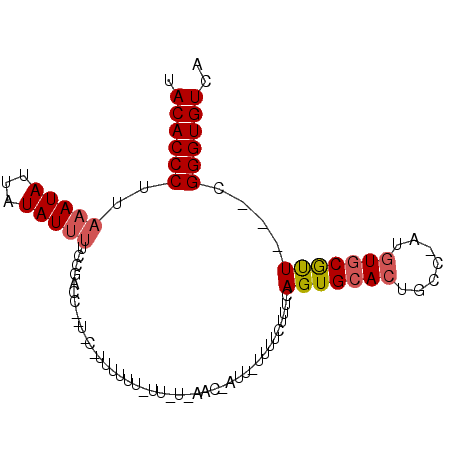
>3L_DroMel_CAF1 21329224 90 - 23771897 UACACCCUUAAAUAUUAUAUUUUCCCUCA--UACAUUUUUUGUUAUUAAC-AUUCUUUUCUUUCAGUGCACUGCC-AUGUGCGUU---CGGGUGUCA .((((((......................--.(((.....))).......-................((((....-..))))...---.)))))).. ( -15.40) >DroSim_CAF1 33216 72 - 1 UACACCCUUAAAUAUUAUAUGAUCCGACC--------------------G-AUUAUUUUCUUUCAGUGCACUGCC-AUGUGCGUU---CGGGUGUCA .((((((...........((((((.....--------------------)-)))))...........((((....-..))))...---.)))))).. ( -17.80) >DroYak_CAF1 58704 92 - 1 UACACCCUUAAAUAUAAUAUUUUCCGCCCAGUUCCUUUUUUUUUUUGAAUU-----UUUCUUUCAGUGGAGUGCCCCUGUGCACUGACAGGGUGUCA .(((((((..............(((((..................((((..-----.....)))))))))((((......))))....))))))).. ( -19.46) >consensus UACACCCUUAAAUAUUAUAUUUUCCGACC__U_C_UUUUUU_UU_U_AAC_AUU_UUUUCUUUCAGUGCACUGCC_AUGUGCGUU___CGGGUGUCA .((((((..(((((...)))))..........................................(((((((.......)))))))....)))))).. ( -9.17 = -10.07 + 0.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:06:23 2006