
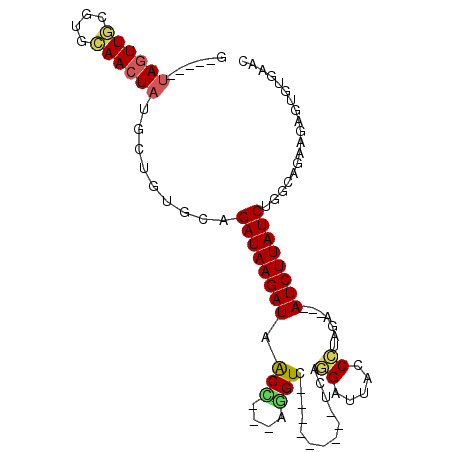
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,324,463 – 21,324,560 |
| Length | 97 |
| Max. P | 0.998031 |

| Location | 21,324,463 – 21,324,560 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 67.45 |
| Mean single sequence MFE | -25.69 |
| Consensus MFE | -10.09 |
| Energy contribution | -10.07 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.39 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21324463 97 + 23771897 A-----UAGUUGCGUGCAACUAUGCUCUGCUGAUAAGAUAACC---AGGUUAGGAUUAGUCCAGGAGAACCCUAGACCUAUCUUAUCUGGCAGCAGAGUGUAAGC (-----((((((....)))))))(((((((((.........((---((((.(((((..(((.(((.....))).)))..))))))))))))))))))))...... ( -38.90) >DroSec_CAF1 27636 85 + 1 G-----UAGUUGUGAGCAACUAUGCUGUGCUGAUAAGAUAACC---AGGUC---------GCAGGAUUACCCUAGA---AUCUUAUCUGGCAGGAGAGUGUAAAC (-----((((((....)))))))(((.(.(((...((((((..---...((---------..(((.....))).))---...))))))..))).).)))...... ( -23.10) >DroSim_CAF1 27684 80 + 1 G-----UAGUUGCGAGCAACUAUGCUGUGCUGAUAAGAUAACC---AGGUC---------GCAGGAUUACCCUAGA---AUCUUAUCUGGCCGCAG-----CAAC (-----((((((....)))))))((((((((((((((((....---...((---------..(((.....))).))---)))))))).)).)))))-----)... ( -27.51) >DroEre_CAF1 29574 80 + 1 G-----UAGUUGCGUGCAACUACG-----CAGAUAAGAUAACU---AAGUC---------UCAGGAUCGCCCCAGA---AUCUUAUCUGUCGGAAGAGUCUGAGC (-----((((((....))))))).-----((((((((((....---...((---------(..((....))..)))---))))))))))(((((....))))).. ( -25.11) >DroYak_CAF1 53927 79 + 1 G-----UAGUUGCGUGAAACUAUG-----CAGAUAAGAUAGCU---AGGUC---------UCAGGAUUACC-CUGA---AUCUUAUCUGACAGAAGAGUCUGAAC (-----(((((......)))))).-----((((((((((....---.....---------(((((.....)-))))---)))))))))).((((....))))... ( -23.80) >DroAna_CAF1 46049 88 + 1 CAUUUUGAAUUCUCUGGAACUG-GUUGGGAAGAUAAGAUAACCCAGAAGUC---------UCAAGUUUAUCUGAAA---AUCUUAUCUUUUUG-UGAGUGUG--- ..................(((.-..(..(((((((((((....((((....---------.........))))...---)))))))))))..)-..)))...--- ( -15.72) >consensus G_____UAGUUGCGUGCAACUAUGCUGUGCAGAUAAGAUAACC___AGGUC_________UCAGGAUUACCCUAGA___AUCUUAUCUGGCAGAAGAGUGUGAAC ......((((((....)))))).........((((((((.(((....))).............((.....)).......)))))))).................. (-10.09 = -10.07 + -0.02)


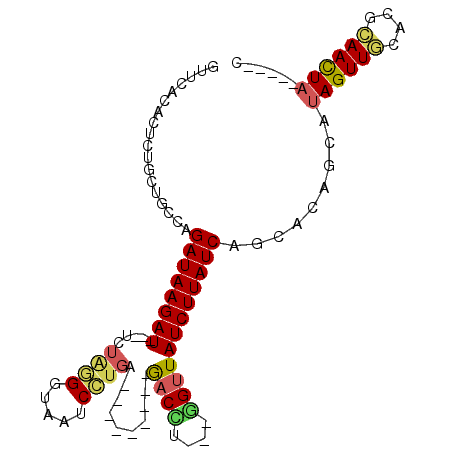
| Location | 21,324,463 – 21,324,560 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 67.45 |
| Mean single sequence MFE | -24.54 |
| Consensus MFE | -12.08 |
| Energy contribution | -12.08 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.49 |
| SVM decision value | 2.99 |
| SVM RNA-class probability | 0.998031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21324463 97 - 23771897 GCUUACACUCUGCUGCCAGAUAAGAUAGGUCUAGGGUUCUCCUGGACUAAUCCUAACCU---GGUUAUCUUAUCAGCAGAGCAUAGUUGCACGCAACUA-----U .......(((((((....(((((((((((((((((.....))))))))...((......---)).)))))))))))))))).(((((((....))))))-----) ( -37.70) >DroSec_CAF1 27636 85 - 1 GUUUACACUCUCCUGCCAGAUAAGAU---UCUAGGGUAAUCCUGC---------GACCU---GGUUAUCUUAUCAGCACAGCAUAGUUGCUCACAACUA-----C .............(((..((((((((---.(((((..........---------..)))---))..)))))))).))).....((((((....))))))-----. ( -21.40) >DroSim_CAF1 27684 80 - 1 GUUG-----CUGCGGCCAGAUAAGAU---UCUAGGGUAAUCCUGC---------GACCU---GGUUAUCUUAUCAGCACAGCAUAGUUGCUCGCAACUA-----C ..((-----(((..((..((((((((---.(((((..........---------..)))---))..)))))))).)).)))))((((((....))))))-----. ( -28.90) >DroEre_CAF1 29574 80 - 1 GCUCAGACUCUUCCGACAGAUAAGAU---UCUGGGGCGAUCCUGA---------GACUU---AGUUAUCUUAUCUG-----CGUAGUUGCACGCAACUA-----C ..............(.((((((((((---.(((((.(........---------).)))---))..))))))))))-----)(((((((....))))))-----) ( -24.90) >DroYak_CAF1 53927 79 - 1 GUUCAGACUCUUCUGUCAGAUAAGAU---UCAG-GGUAAUCCUGA---------GACCU---AGCUAUCUUAUCUG-----CAUAGUUUCACGCAACUA-----C .....((..((..((.((((((((((---((((-(.....)))))---------.....---....))))))))))-----)).))..)).........-----. ( -19.30) >DroAna_CAF1 46049 88 - 1 ---CACACUCA-CAAAAAGAUAAGAU---UUUCAGAUAAACUUGA---------GACUUCUGGGUUAUCUUAUCUUCCCAAC-CAGUUCCAGAGAAUUCAAAAUG ---........-....((((((((((---.((((((.........---------....))))))..))))))))))......-.(((((....)))))....... ( -15.02) >consensus GUUCACACUCUGCUGCCAGAUAAGAU___UCUAGGGUAAUCCUGA_________GACCU___GGUUAUCUUAUCAGCACAGCAUAGUUGCACGCAACUA_____C ..................((((((((.....((((.....))))..........((((....)))))))))))).........((((((....))))))...... (-12.08 = -12.08 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:06:21 2006