
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,318,773 – 21,318,980 |
| Length | 207 |
| Max. P | 0.999315 |

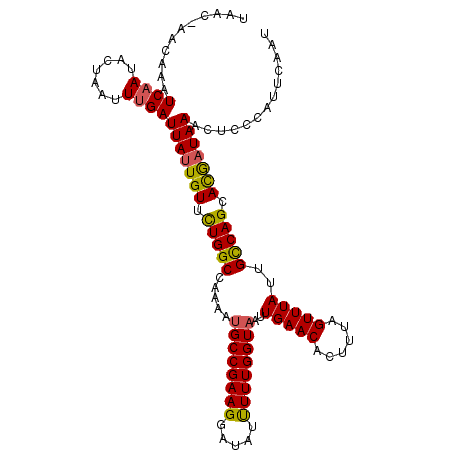
| Location | 21,318,773 – 21,318,883 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 93.77 |
| Mean single sequence MFE | -22.50 |
| Consensus MFE | -20.64 |
| Energy contribution | -20.28 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.51 |
| SVM RNA-class probability | 0.999315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21318773 110 - 23771897 UAAC-AACAAAUCAAUACUAAUUUGAUUAUUGUUCUGGCCAAAAUGCCGAAGGAUAUUUUUGGUAAUUGAACACUUUAGUUUAUUGCCAGCACGCUAAACUCCCAUUCAAU ....-....((((((.......))))))...((.(((((.....((((((((.....))))))))..(((((......)))))..))))).)).................. ( -20.80) >DroSec_CAF1 21925 111 - 1 UAACCAACAAAUCAAUACUAAUUUGAUUAUUGUUCUGGCCAAAAUGCCGAAGGAUAUUUUUGGUAAUUGAACACUUUAGUUUAUUGCCAGCACGAUAAACUCCCAUUCAAU ..(((((..((((((.......))))))(((.(((.(((......)))))).)))....))))).((((((......(((((((((......)))))))))....)))))) ( -24.10) >DroSim_CAF1 21971 111 - 1 UAACCAACAAAUCAAUACUAAUUUGAUUAUUGUUCUGGCCAAAAUGCCGAAGGAUAUUUUUGGUAAUUGAACACUUUAGUUUAUUGCCAGCACGAUAAACUCCCAUUCAAU ..(((((..((((((.......))))))(((.(((.(((......)))))).)))....))))).((((((......(((((((((......)))))))))....)))))) ( -24.10) >DroEre_CAF1 23838 110 - 1 UAAC-AAUAAAUCAAUACUAAUUCGAUUAUUGUUUUGGCCAAAAUGCCGAAGGAUAUUUUUGGUAAUUGAACACUUUAGUUUAUUGUCAGCAUAAUAAACUCCUAUUCAAU ....-......((((((((((...(((...(.(((((((......))))))).).))).))))).))))).......((((((((((....)))))))))).......... ( -19.90) >DroYak_CAF1 48089 110 - 1 UAAC-AAGAAAUCAAUACUAAUUUGAUUAUUGUUCUGGCCAAAAGGCCGAAGGAAAUCUUUGGUAAUUGAACACUUUAGUUUAUUGUCAGCAUGAUAAUCUCGCACUCAAU ....-..((..((((((((((...((((.((.(((.((((....))))))).)))))).))))).)))))........((..((((((.....))))))...))..))... ( -23.60) >consensus UAAC_AACAAAUCAAUACUAAUUUGAUUAUUGUUCUGGCCAAAAUGCCGAAGGAUAUUUUUGGUAAUUGAACACUUUAGUUUAUUGCCAGCACGAUAAACUCCCAUUCAAU ...........((((.......))))(((((((.(((((.....((((((((.....))))))))..(((((......)))))..))))).)))))))............. (-20.64 = -20.28 + -0.36)


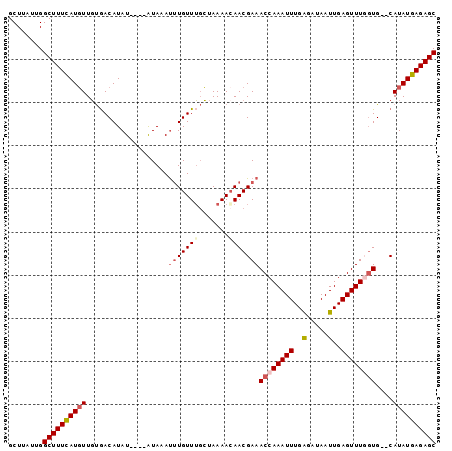
| Location | 21,318,883 – 21,318,980 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -21.68 |
| Consensus MFE | -15.95 |
| Energy contribution | -17.45 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.702317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21318883 97 + 23771897 GCUUAUUGGCUUUUAUGUUGUGACAUAUGUAUGUAAAUUUGUUUGCUAAAACAACGAAACCAAAUUUGAGAUAAUUGAGUUUUGUG--CAUAUGAGAGC ........((((((((((.((.((((....)))).....(((((....)))))(((((((..((((......))))..))))))))--))))))))))) ( -19.50) >DroSec_CAF1 22036 93 + 1 GCUUAUUGGCUUUCAUGUUGUGACAUAU----AUAAAUGUGUUUGCUAAAACAACGAAACCAAAUUUGAGAUAAUUGAGUUUGCUG--CAUAUGAGAGC ........((((((((((((((((((((----....))))))).......))))).....((((((..(.....)..))))))...--...)))))))) ( -21.31) >DroSim_CAF1 22082 95 + 1 GCUUAUUGGCUUUCAUGUUGUGACAUAU----AUAAAUUUGUUUGCUAAAAAAACGAAACCAAAUUUGAGAUAAUUGAGUUUGGUGCGCAUAUGAGAGC ........((((((((((.(((.(....----.....(((((((.......)))))))((((((((..(.....)..)))))))))))))))))))))) ( -28.30) >DroYak_CAF1 48199 93 + 1 GCUUAUUGGCUUUCAUUUUCCAACAUAU----AUAAACUUGUUGCGUAAAACAACGAAACCAAAUUUGGGAUAAUUGAGUUUUGUG--CAUAUGAGAGC ........((((((((....(((((...----.......))))).........(((((((..((((......))))..))))))).--...)))))))) ( -17.60) >consensus GCUUAUUGGCUUUCAUGUUGUGACAUAU____AUAAAUUUGUUUGCUAAAACAACGAAACCAAAUUUGAGAUAAUUGAGUUUGGUG__CAUAUGAGAGC ........((((((((((...................(((((((.......)))))))((((((((..(.....)..))))))))....)))))))))) (-15.95 = -17.45 + 1.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:06:19 2006