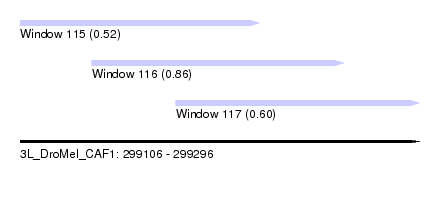
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 299,106 – 299,296 |
| Length | 190 |
| Max. P | 0.858785 |

| Location | 299,106 – 299,220 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 86.11 |
| Mean single sequence MFE | -30.05 |
| Consensus MFE | -20.94 |
| Energy contribution | -21.00 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 299106 114 + 23771897 AUCAACUCCAUAAUAAACCUUUUGUUGCUGCACCUGCAGUGAAAUCGCGAAUCUUUCAAGUAGUUGUUCGCCUAGUCGCAUUCUCAGCAUUUUUCUGCCAGCGGGCAGUUAGCA ......................(((((((((.((.((.((((....((((((((.......))..))))))....)))).......(((......)))..)))))))).))))) ( -26.20) >DroSec_CAF1 9024 114 + 1 AUUAACUGCAUAAUAAACCUUUUGUUGCUGCACCUGCAGUUAAGUCGCGAAUCUUUCAAGUGGUUGUUCGCUUAGUCGCAUUCUCAGCAUAUUUCUGCUGACAGGCAGGCUGCA ......((((((((((.....)))))).))))...((((((..(((((((.......((((((....))))))..))))....((((((......))))))..))).)))))). ( -31.60) >DroSim_CAF1 11355 114 + 1 AUUAACUGCAUAAUAAACCUUUUGUUGCUGCACCUGCAGUUAAGUCGCGAAUCUUUCAAGUGGUUGUUCGCUUAGUCGCAUUCUCAGCAUAUUUCUGCUGACAGGCAGGCUGCA ......((((((((((.....)))))).))))...((((((..(((((((.......((((((....))))))..))))....((((((......))))))..))).)))))). ( -31.60) >DroYak_CAF1 9950 106 + 1 AUUAACUGCAUAGUAAACCUUUUGUGGCUGCACCUGCAGUCAACUCGCGAAUCUUUCAGGUGGUUG--------UUCGCAUUCUCAGCAUAUUUCUGCUUGGAGGCGGACUGUA ...(((..(..((...........(((((((....)))))))..........)).....)..)))(--------(((((.((((.((((......)))).)))))))))).... ( -30.80) >consensus AUUAACUGCAUAAUAAACCUUUUGUUGCUGCACCUGCAGUUAAGUCGCGAAUCUUUCAAGUGGUUGUUCGCUUAGUCGCAUUCUCAGCAUAUUUCUGCUGACAGGCAGGCUGCA ..(((((((...............(((((((....)))))))......((.....))..)))))))....................(((....((((((....)))))).))). (-20.94 = -21.00 + 0.06)

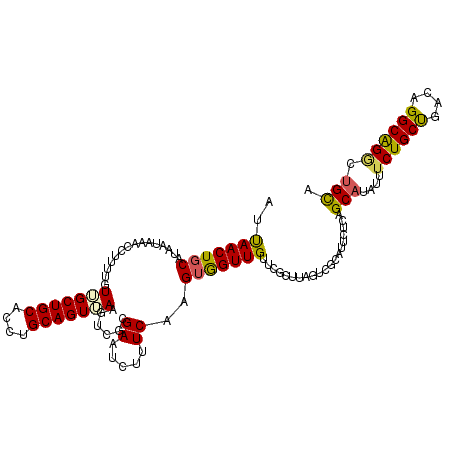
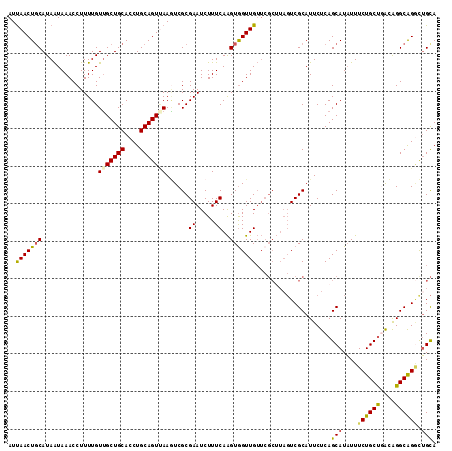
| Location | 299,140 – 299,260 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.39 |
| Mean single sequence MFE | -35.00 |
| Consensus MFE | -18.27 |
| Energy contribution | -20.34 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 299140 120 + 23771897 UGCAGUGAAAUCGCGAAUCUUUCAAGUAGUUGUUCGCCUAGUCGCAUUCUCAGCAUUUUUCUGCCAGCGGGCAGUUAGCAUCGUCAGUUUUUGCGUAUUCGUAAAAAUUGAACAGUGCAC ....((((....((((((((.......))..))))))....))))...............(((((....)))))...((((..((((((((((((....))))))))))))...)))).. ( -36.00) >DroSec_CAF1 9058 120 + 1 UGCAGUUAAGUCGCGAAUCUUUCAAGUGGUUGUUCGCUUAGUCGCAUUCUCAGCAUAUUUCUGCUGACAGGCAGGCUGCAUCAUCAAUUGUUGCGUAUUCGUAAAAAUUGAACAGUGCAC (((((((..(((((((.......((((((....))))))..))))....((((((......))))))..))).)))))))...((((((.(((((....))))).))))))......... ( -35.90) >DroSim_CAF1 11389 120 + 1 UGCAGUUAAGUCGCGAAUCUUUCAAGUGGUUGUUCGCUUAGUCGCAUUCUCAGCAUAUUUCUGCUGACAGGCAGGCUGCAUUGUCAAUUGUUGCGUAUUCGUAAAAAUUGAACAGUGCAC ........((((((((((...((....))..))))))......((....((((((......))))))...)).))))((((((((((((.(((((....))))).))))).))))))).. ( -38.80) >DroYak_CAF1 9984 90 + 1 UGCAGUCAACUCGCGAAUCUUUCAGGUGGUUG--------UUCGCAUUCUCAGCAUAUUUCUGCUUGGAGGCGGACUGUAUCGCCAAUUGUUGCGUUC---------------------- .((((.(((.....((.....)).((((((.(--------(((((.((((.((((......)))).))))))))))...))))))..)))))))....---------------------- ( -29.30) >consensus UGCAGUUAAGUCGCGAAUCUUUCAAGUGGUUGUUCGCUUAGUCGCAUUCUCAGCAUAUUUCUGCUGACAGGCAGGCUGCAUCGUCAAUUGUUGCGUAUUCGUAAAAAUUGAACAGUGCAC ((((((......((((......(((....))).........))))...............(((((....)))))))))))...((((((.(((((....))))).))))))......... (-18.27 = -20.34 + 2.06)

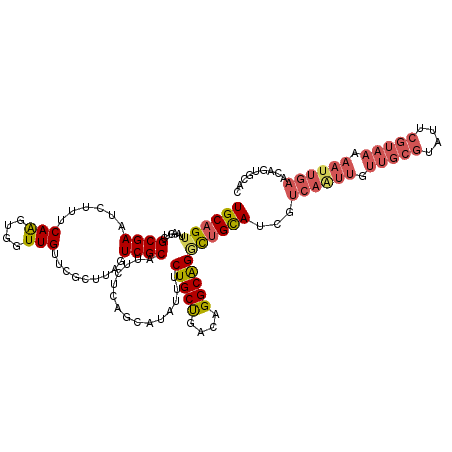
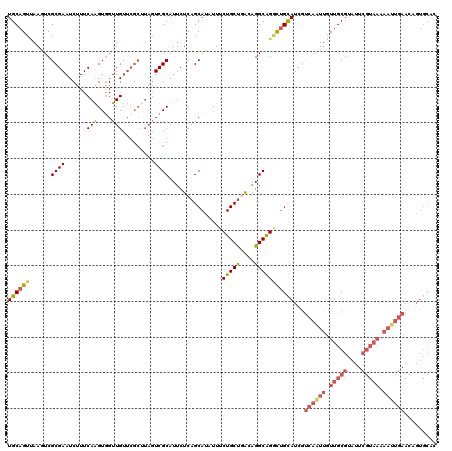
| Location | 299,180 – 299,296 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 90.80 |
| Mean single sequence MFE | -32.03 |
| Consensus MFE | -23.34 |
| Energy contribution | -23.23 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598650 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 299180 116 + 23771897 GUCGCAUUCUCAGCAUUUUUCUGCCAGCGGGCAGUUAGCAUCGUCAGUUUUUGCGUAUUCGUAAAAAUUGAACAGUGCACCACGUCGAUUCUUAACAUGAAGUUUCUGUUUAACUU (((((...............(((((....)))))...((((..((((((((((((....))))))))))))...)))).....).))))..........(((((.......))))) ( -29.20) >DroSec_CAF1 9098 116 + 1 GUCGCAUUCUCAGCAUAUUUCUGCUGACAGGCAGGCUGCAUCAUCAAUUGUUGCGUAUUCGUAAAAAUUGAACAGUGCACCACAUCGAUUCUUAACAUGCAGUUUCUGUUUAACUU .........((((((......)))))).(((((((((((((..((((((.(((((....))))).))))))...(((...))).............)))))))..))))))..... ( -30.20) >DroSim_CAF1 11429 116 + 1 GUCGCAUUCUCAGCAUAUUUCUGCUGACAGGCAGGCUGCAUUGUCAAUUGUUGCGUAUUCGUAAAAAUUGAACAGUGCACCACAUCGAUUCUUAUCAUGCAGUUUCUGUGUAACUU ((..(((..((((((......))))))...(((((.(((((((((((((.(((((....))))).))))).)))))))))).....(((....))).))).......)))..)).. ( -36.70) >consensus GUCGCAUUCUCAGCAUAUUUCUGCUGACAGGCAGGCUGCAUCGUCAAUUGUUGCGUAUUCGUAAAAAUUGAACAGUGCACCACAUCGAUUCUUAACAUGCAGUUUCUGUUUAACUU ...(((....(.((((....(((((....)))))...((((..((((((.(((((....))))).))))))...))))..................)))).)....)))....... (-23.34 = -23.23 + -0.11)

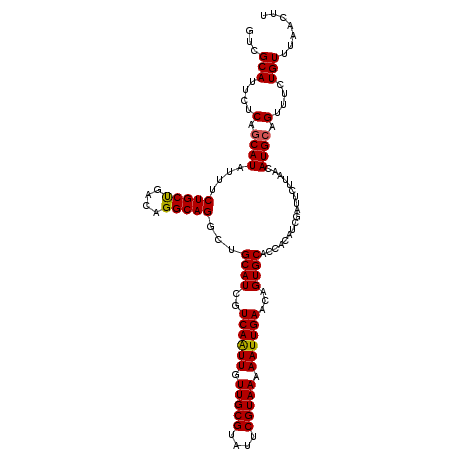
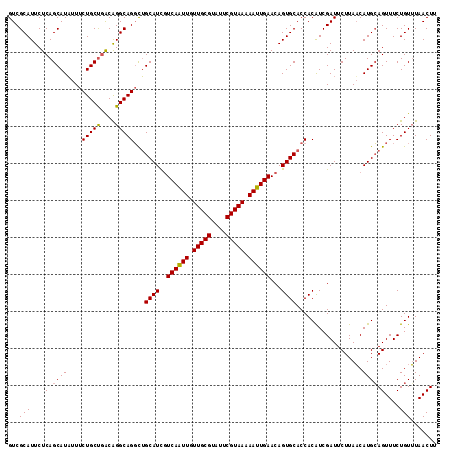
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:52 2006