
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,315,248 – 21,315,392 |
| Length | 144 |
| Max. P | 0.853498 |

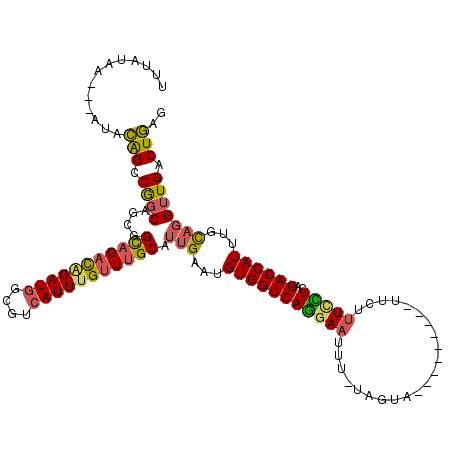
| Location | 21,315,248 – 21,315,355 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.85 |
| Mean single sequence MFE | -34.73 |
| Consensus MFE | -26.92 |
| Energy contribution | -26.73 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
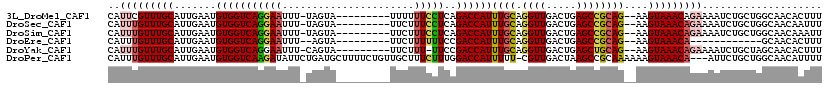
>3L_DroMel_CAF1 21315248 107 + 23771897 UUUAUAA---AUACAGCCGGCAGCGGCAGACAGAUGGCGUCAUUCGUUUGCAUUGAAUGUGGUCAGGAAUUU-UAGUA---------UUUUUUCCUCAGACCAUUUGCAGGUUGACUGAG .......---...(((((.((((..((((((..(((....)))..)))))).......(((((((((((...-.....---------....)))))..)))))))))).)))))...... ( -32.90) >DroSec_CAF1 18361 107 + 1 UUUAUAA---AUACAGCCGGCAGCGGCAGACAGAUGGCGUCAUUUGUUUGCAUUGAAUGUGGUCAGGAAUUU-UAGUA---------UUCUUUCCUCAGACCAUUUGCAGGUUGACUGAG .......---...(((((.((((..(((((((((((....))))))))))).......(((((((((((...-.....---------....)))))..)))))))))).)))))...... ( -36.50) >DroSim_CAF1 18432 107 + 1 UUUAUAA---AUACAGCCGGCAGCGGCAGACAGAUGGCGUCAUUUGUUUGCAUUGAAUGUGGUCAGGAAUUU-UAGUA---------UUCUUUCCUCAGACCAUUUGCAGGUUGACUGAG .......---...(((((.((((..(((((((((((....))))))))))).......(((((((((((...-.....---------....)))))..)))))))))).)))))...... ( -36.50) >DroEre_CAF1 20365 106 + 1 UUUAUAA---AUACGGUCGGCAGAGGCAGACAGAUGGCGUCAUUUGUUUGCAUUGAAUGUGGUCAGGAAUUU--AGUA---------UUCUUUUUUCCGACCAUUUGCAGGUUGACUGAG .......---...((((((((....(((((((((((....))))))))))).(((...((((((.((((...--((..---------..))...))))))))))...))))))))))).. ( -40.30) >DroYak_CAF1 44583 106 + 1 UUUAUAA---AUACGGUCGCCAGCGGCAGACAGAUGGCGUCAUUUGUUUGCAUUGAAUGUGGUCAGGAAUUU-CAGUA---------UUCUUU-UUCCGACCAUUUGCAGGUUGACUGAG .......---...((((((((.((((((((((((((....))))))))))).......((((((.((((...-.((..---------..))..-)))))))))).))).)).)))))).. ( -37.50) >DroPer_CAF1 34381 118 + 1 UUUAUAAAAAAUAUGGCCAGCAGACGUUG-UGGAUUGCGUCAUUUGUUUGCAUUGAAUGUGGUCAAGAUAUUCUGAUGCUUUUCUGUUGCUUUCUUUGGACCAUUUUU-CGUUGACUAAG ........(((.((((((((.(((.((.(-..((..((((((((((.(..(((...)))..).))))......))))))...))..).))..))))))).)))).)))-........... ( -24.70) >consensus UUUAUAA___AUACAGCCGGCAGCGGCAGACAGAUGGCGUCAUUUGUUUGCAUUGAAUGUGGUCAGGAAUUU_UAGUA_________UUCUUUCCUCAGACCAUUUGCAGGUUGACUGAG .............(((.((((....(((((((((((....))))))))))).(((...(((((((((((......................)))))..))))))...))))))).))).. (-26.92 = -26.73 + -0.19)


| Location | 21,315,285 – 21,315,392 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 83.39 |
| Mean single sequence MFE | -27.79 |
| Consensus MFE | -18.82 |
| Energy contribution | -19.43 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21315285 107 + 23771897 CAUUCGUUUGCAUUGAAUGUGGUCAGGAAUUU-UAGUA---------UUUUUUCCUCAGACCAUUUGCAGGUUGACUGAGCCGCAG--AAGUAAACAGAAAAUCUGCUGGCAACACUUU .....(((.((.(((...(((((((((((...-.....---------....)))))..))))))...))))).)))...(((((((--(.............))))).)))........ ( -27.82) >DroSec_CAF1 18398 107 + 1 CAUUUGUUUGCAUUGAAUGUGGUCAGGAAUUU-UAGUA---------UUCUUUCCUCAGACCAUUUGCAGGUUGACUGAGCCGCAG--AAGUAAACAGAAAAUCUGCUGGCAACAAUUU ..(((((((((........((((((((((...-.....---------....)))))..)))))(((((.((((.....))))))))--).)))))))))........((....)).... ( -29.40) >DroSim_CAF1 18469 107 + 1 CAUUUGUUUGCAUUGAAUGUGGUCAGGAAUUU-UAGUA---------UUCUUUCCUCAGACCAUUUGCAGGUUGACUGAGCCGCAG--AAGUAAACAGAAAAUCUGCUGGCAACAAAUU ..(((((((((........((((((((((...-.....---------....)))))..)))))(((((.((((.....))))))))--).)))))))))........((....)).... ( -29.40) >DroEre_CAF1 20402 94 + 1 CAUUUGUUUGCAUUGAAUGUGGUCAGGAAUUU--AGUA---------UUCUUUUUUCCGACCAUUUGCAGGUUGACUGAGCCGCAG--AAGUAAACA------------GCAACACUUU ...((((((((........(((((.((((...--((..---------..))...)))))))))(((((.((((.....))))))))--).)))))))------------)......... ( -27.30) >DroYak_CAF1 44620 106 + 1 CAUUUGUUUGCAUUGAAUGUGGUCAGGAAUUU-CAGUA---------UUCUUU-UUCCGACCAUUUGCAGGUUGACUGAGCUGCAG--AAGUAAACAGAAAAUCUGCUAGCAACACUUU ..(((((((((........(((((.((((...-.((..---------..))..-)))))))))(((((((.((....)).))))))--).))))))))).................... ( -28.80) >DroPer_CAF1 34420 115 + 1 CAUUUGUUUGCAUUGAAUGUGGUCAAGAUAUUCUGAUGCUUUUCUGUUGCUUUCUUUGGACCAUUUUU-CGUUGACUAAGCCGCAAAAAAGUAAACA---AUUCUGCUGGCAACAUUUU ...((((..(((..((((((.......))))))...(((((((.(((.((((((..((((......))-))..))..)))).))).)))))))....---....)))..))))...... ( -24.00) >consensus CAUUUGUUUGCAUUGAAUGUGGUCAGGAAUUU_UAGUA_________UUCUUUCCUCAGACCAUUUGCAGGUUGACUGAGCCGCAG__AAGUAAACAGAAAAUCUGCUGGCAACACUUU ..(((((((((.......(((((((((((......................)))))..))))))((((.((((.....))))))))....))))))))).................... (-18.82 = -19.43 + 0.62)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:06:17 2006