
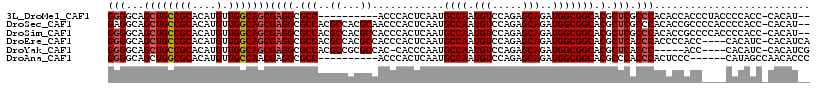
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,314,108 – 21,314,212 |
| Length | 104 |
| Max. P | 0.573807 |

| Location | 21,314,108 – 21,314,212 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 83.58 |
| Mean single sequence MFE | -38.45 |
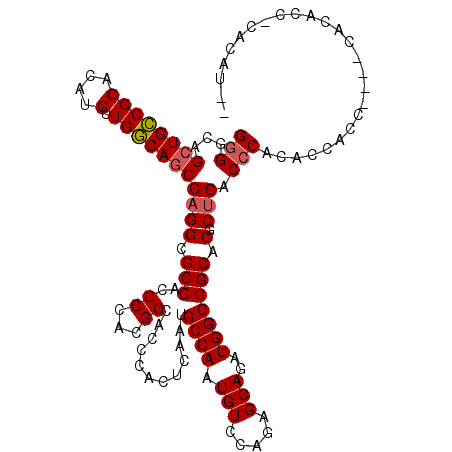
| Consensus MFE | -28.66 |
| Energy contribution | -29.05 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21314108 104 + 23771897 GGGGCAGCUGCCGCACAUGUUGGCAGCGAGGCGCC----------ACCCACUCAAUGCCAAUGUCCAGAGCAGAUGGCGGCACGCUCGCCCACACCACCCUACCCCACC-CACAU-- .(((((((((((((.((((((((((..(((.....----------.....)))..)))))))((.....))..))))))))).))).))))..................-.....-- ( -38.80) >DroSec_CAF1 17153 114 + 1 GAGGCAGCUGCCGCACAUGUUGGCAGCGAGGCGCCACGCCACGCAACCCACUCAAUGCCAAUGUCCAGAGCAGAUGGCGGCACGCUCGCCCACACCGCCCCACCCCACC-CACAU-- (.((((((((((((.(((((((((((((.((((...)))).)))...........)))))))((.....))..))))))))).))).))))..................-.....-- ( -39.00) >DroSim_CAF1 17220 114 + 1 GGGGCAGCUGCCGCACAUGUUGGCAGCGAGGCGCCACGCCACGCCACCCACUCAAUGCCAAUGUCCAGAGCAGAUGGCGGCACGCUCGCCCACACCGCCCCACCCCACC-CACAU-- (((((.((((((((....).)))))))(.((((...((...(((((.(..(((...((....))...)))..).)))))...))..))))).....)))))........-.....-- ( -43.30) >DroEre_CAF1 19114 112 + 1 GGGGCAGCUGCCGCACAUGUUGGCAGCGAGGCGCCACGCCACGCCACCCACUCAAUGCCAAUGUCCAGAGCAGAUGGCGGCACGCUCACCCACCCCACC----CACAUC-CACAUCA (((..(((((((((.(((((((((((((.((((...)))).)))...........)))))))((.....))..))))))))).)))..)))........----......-....... ( -40.00) >DroYak_CAF1 43407 106 + 1 GGGGCAGCUGCCGCACAUGUUGGCAGCGAGGCGCCACGCCGCGCCAC-CACCCAAUGCCAAUGUCCAGAGCAGAUGGCGGCACGCUCACCC-----ACC----CACAUC-CACAUCG (((..(((((((((.(((((((((((((.((((...)))).)))...-.......)))))))((.....))..))))))))).)))..)))-----...----......-....... ( -40.60) >DroAna_CAF1 35257 101 + 1 GGGGCAGCUGGCGCACAUGUUGCCAACGAGGCGCC----------ACCCACUCAAUGCCAAUGUCCAGAGCAGAUGGCGGCACGCCCACCCACUCCC------CAUAGCCAACACCC .((((.((((((((.(.(((.....))).))))))----------...........((((.(((.....)))..)))))))..))))..........------.............. ( -29.00) >consensus GGGGCAGCUGCCGCACAUGUUGGCAGCGAGGCGCCACGCCACGCCACCCACUCAAUGCCAAUGUCCAGAGCAGAUGGCGGCACGCUCACCCACACCACC____CACACC_CACAU__ (((...((((((((....).)))))))((((.(((..((...))............((((.(((.....)))..))))))).).))).))).......................... (-28.66 = -29.05 + 0.39)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:06:15 2006