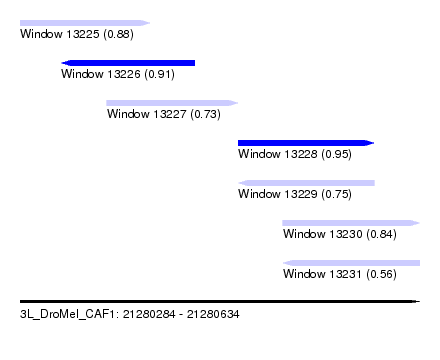
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,280,284 – 21,280,634 |
| Length | 350 |
| Max. P | 0.951425 |

| Location | 21,280,284 – 21,280,398 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.56 |
| Mean single sequence MFE | -32.32 |
| Consensus MFE | -22.85 |
| Energy contribution | -21.73 |
| Covariance contribution | -1.13 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21280284 114 + 23771897 UUUCGAUCAUGCAAAGGGUG----UGCAUGUAAAGGACCUUGGUUGCAAGCCACGCAGCUCAAGGGUAAAUGAAUGUACGAUAUGUGUGCGGUAUAC--AUGUACAUAUGUAUGCAUAUG ........(((((....(((----((((((((.....((((((((((.......))))).))))).........((((((.....))))))...)))--)))))))).....)))))... ( -37.70) >DroSec_CAF1 38554 108 + 1 UUUUGAUCAUGCAAAGGGUG----UGCAUGUAAAGGACCUUGGUGGCAAGCCACGCAGCUCAAGGGUAAAUGAAUGUACGAUAUGCGUGCG--------AUGUACAUACGUAUGUAUAUG .......((((((.(...((----(((((........(((((((((....))))......)))))........))))))).).))))))..--------((((((((....)))))))). ( -30.19) >DroSim_CAF1 40805 108 + 1 UUUUGAUCAUGCAAAGGGUG----UGCAUGUAAAGGACCUUGGUGGCAAGCCACGCAGCUCAGGGGUAAAUGAAUGUACGAUAUGCGUGCG--------AUGUACAUACGUAUGUAUAUG .......((((((.(...((----(((((........(((((((((....))))......)))))........))))))).).))))))..--------((((((((....)))))))). ( -29.89) >DroEre_CAF1 39750 119 + 1 ACUCGAUCACCUACAUGGUGUAUGUACGUGUAAAGGUCCUUGAUUGCAAUCCACACUGUUCAAGGGUAGAUGAAUGUACGAUAUGCGUACA-UAAACAUACAUACAUCCAUAUGUAUAUG .......((((.....))))(((((((((((......((((((..(((........)))))))))(((.(((.(((((((.....))))))-)...)))...)))....))))))))))) ( -31.50) >consensus UUUCGAUCAUGCAAAGGGUG____UGCAUGUAAAGGACCUUGGUGGCAAGCCACGCAGCUCAAGGGUAAAUGAAUGUACGAUAUGCGUGCG________AUGUACAUACGUAUGUAUAUG ((((...((((((...........))))))...))))(((((((.((.......)).)).))))).........((((((.....))))))........((((((((....)))))))). (-22.85 = -21.73 + -1.13)

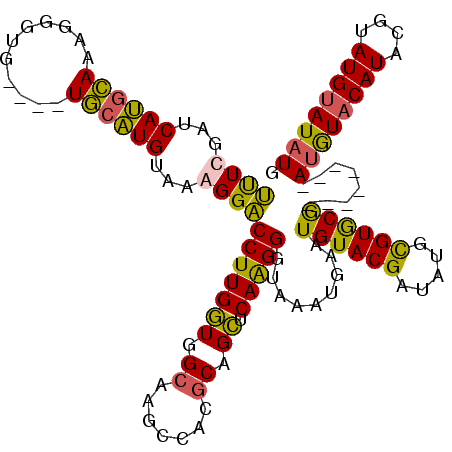
| Location | 21,280,320 – 21,280,437 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.71 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -22.05 |
| Energy contribution | -22.68 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21280320 117 - 23771897 GUGGACACGCGGCAGUUACACUAA-ACACGUGUACACAAGCAUAUGCAUACAUAUGUACAU--GUAUACCGCACACAUAUCGUACAUUCAUUUACCCUUGAGCUGCGUGGCUUGCAACCA (..(.((((((((...........-....(((((((...(((((((....)))))))...)--))))))..................(((........)))))))))))..)..)..... ( -32.60) >DroSec_CAF1 38590 112 - 1 GUGGACACGCGGCAGUCACACCAAUACAUGUGUACCAAAGCAUAUACAUACGUAUGUACAU--------CGCACGCAUAUCGUACAUUCAUUUACCCUUGAGCUGCGUGGCUUGCCACCA ((((.((((((((..............((((((......))))))...(((((((((....--------.....))))).))))...(((........))))))))))).....)))).. ( -30.20) >DroSim_CAF1 40841 112 - 1 GUGGACACGCGGCAGUCACACCAAUACAUGUGUACCUAAGCAUAUACAUACGUAUGUACAU--------CGCACGCAUAUCGUACAUUCAUUUACCCCUGAGCUGCGUGGCUUGCCACCA ((((.((((((((..............((((((......))))))...(((((((((....--------.....))))).)))).................)))))))).....)))).. ( -30.10) >DroEre_CAF1 39790 118 - 1 GUGGACACGCGGCU-UCACACCAAUGCAUUUGUACACAAGCAUAUACAUAUGGAUGUAUGUAUGUUUA-UGUACGCAUAUCGUACAUUCAUCUACCCUUGAACAGUGUGGAUUGCAAUCA ........((((.(-((((((..((((..(((....))))))).......((((((.(((((((..((-((....)))).))))))).))))))..........)))))))))))..... ( -32.30) >consensus GUGGACACGCGGCAGUCACACCAAUACAUGUGUACACAAGCAUAUACAUACGUAUGUACAU________CGCACGCAUAUCGUACAUUCAUUUACCCUUGAGCUGCGUGGCUUGCAACCA (..(.((((((((................((((((....((((((......)))))).............(....).....))))))(((........)))))))))))..)..)..... (-22.05 = -22.68 + 0.63)

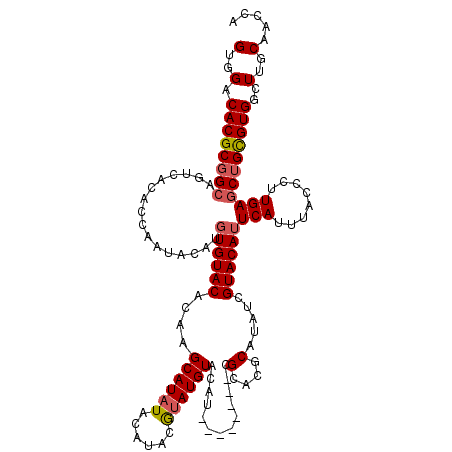
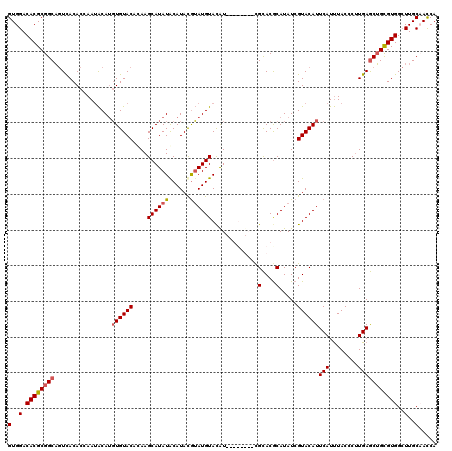
| Location | 21,280,360 – 21,280,475 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.66 |
| Mean single sequence MFE | -33.88 |
| Consensus MFE | -24.54 |
| Energy contribution | -24.23 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21280360 115 + 23771897 AUAUGUGUGCGGUAUAC--AUGUACAUAUGUAUGCAUAUGCUUGUGUACACGUGU-UUAGUGUAACUGCCGCGUGUCCACUCGAGACACGUUAUAAAAUCCACCCUCAGCGGGUC--GCU ....(((.(((((.(((--((..(((..((((((((......))))))))..)))-...)))))))))))))((((((....).)))))............((((.....)))).--... ( -38.40) >DroSec_CAF1 38630 112 + 1 AUAUGCGUGCG--------AUGUACAUACGUAUGUAUAUGCUUUGGUACACAUGUAUUGGUGUGACUGCCGCGUGUCCACUCGAGGCACGUUAUAAAAUCCACCCUCAGAGGGCCACACU (((((((((((--------((....)).)))))))))))((..((((.(((((......)))))))))..))(((.((.(((((((...(((....)))....)))).))))).)))... ( -33.20) >DroSim_CAF1 40881 112 + 1 AUAUGCGUGCG--------AUGUACAUACGUAUGUAUAUGCUUAGGUACACAUGUAUUGGUGUGACUGCCGCGUGUCCACUCGAGGCACGUUAUAAAAUCCACCCUCAGAGGGCCACACU (((((((((((--------((....)).)))))))))))((...(((.(((((......))))))))...))(((.((.(((((((...(((....)))....)))).))))).)))... ( -33.70) >DroEre_CAF1 39830 114 + 1 AUAUGCGUACA-UAAACAUACAUACAUCCAUAUGUAUAUGCUUGUGUACAAAUGCAUUGGUGUGA-AGCCGCGUGUCCACUCGAGGCACGUUAUAAAAUCCACCCUCAGAGGGC----CU ..(((((((((-(((.((((..(((((....))))))))).)))))))).........(((....-.)))))))((((.((.((((...(((....)))....)))))).))))----.. ( -30.20) >consensus AUAUGCGUGCG________AUGUACAUACGUAUGUAUAUGCUUGGGUACACAUGUAUUGGUGUGACUGCCGCGUGUCCACUCGAGGCACGUUAUAAAAUCCACCCUCAGAGGGCC__ACU (((((((((((.................)))))))))))((.....(((((........)))))...)).((((((((....).)))))))...........(((.....)))....... (-24.54 = -24.23 + -0.31)

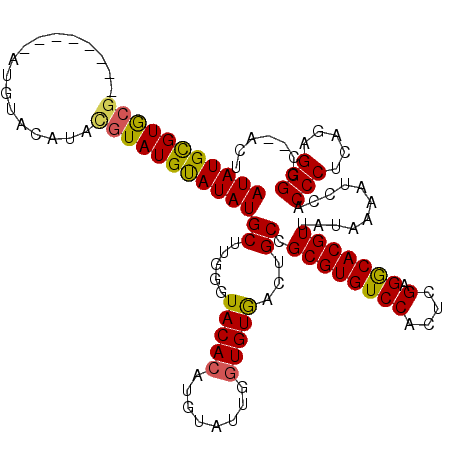
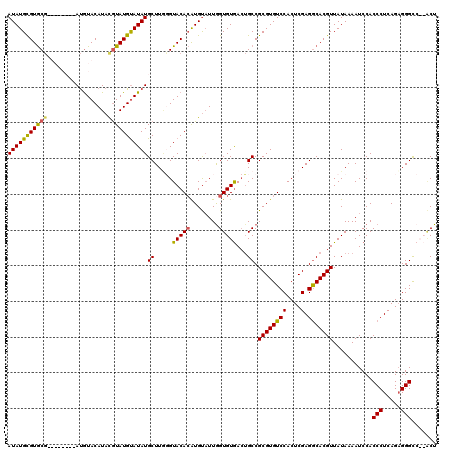
| Location | 21,280,475 – 21,280,594 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.93 |
| Mean single sequence MFE | -42.22 |
| Consensus MFE | -35.90 |
| Energy contribution | -36.27 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951425 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21280475 119 + 23771897 UAAGGGCCAAGUUAACAGACAUAUCUGGUCC-CUUGUCCCUUUGCGACUACUUACCCCUUUGGUGUUGUUGUUGUUGUUGCGCCGCAACGUUGUUGUUGGCAGGGCGGCAAUAGCAGCCA .(((((.((((....((((....))))....-)))).)))))..........((((.....))))..((((((((((((((.((.(((((....)))))...)))))))))))))))).. ( -41.60) >DroSec_CAF1 38742 120 + 1 UAAAGGCCAAGUUAACAGACAUAUCUGGUCCCCUUUACCCCUUGCGACAACUUACCCCUUUGGUGUUGCUGUUGUUGUUGCGACGCAACGUUGUUGUUGGCAAGGCGGCAAUUGCAGCCA ....(((..((((..((((....))))..........((((((((((((((..(((.....)))(((((.(((((....))))))))))...)))))).)))))).)).))))...))). ( -39.40) >DroSim_CAF1 40993 120 + 1 UAAAGGCCAAGUUAACAGACAUAUCUGGUCCCCUUGUCCCCUUGCGACAACUUACCCCUUUGGUGUUGCUGUUGUUGUUGCGACGCAACGUUGUUGUUGGCAAGGCGGCAAUUGCAGCCA ....(((..((((..((((....))))(((.(((((((.....((((((((..(((.....)))(((((.(((((....)))))))))))))))))).))))))).)))))))...))). ( -44.10) >DroEre_CAF1 39944 114 + 1 UAAAGGCCAAGUUAGCAGACAUAUCUGGC------UUCCCCUUGCGACAACUUACCCCUUUGGUGUUGCUGUUGCUGUUGCGCCGCAACGUUGUUGUUGGCAAGGCGGCAAUAGCAGCCA ((((((..(((((..((((....))))((------........))...)))))...))))))(.(((((((((((((((..((((((((...)))).))))..)))))))))))))))). ( -43.80) >consensus UAAAGGCCAAGUUAACAGACAUAUCUGGUCC_CUUGUCCCCUUGCGACAACUUACCCCUUUGGUGUUGCUGUUGUUGUUGCGACGCAACGUUGUUGUUGGCAAGGCGGCAAUAGCAGCCA .....((((......((((....))))................((((((((..(((.....)))(((((.(((((....))))))))))))))))))))))..(((.((....)).))). (-35.90 = -36.27 + 0.37)

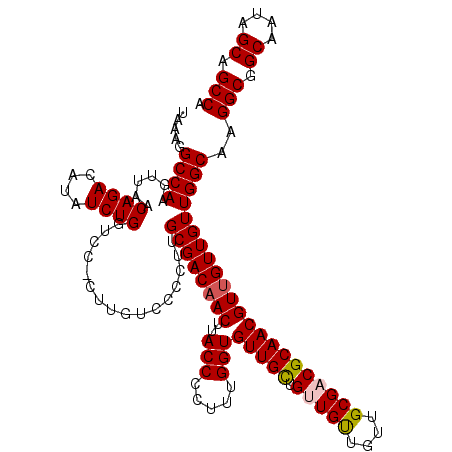
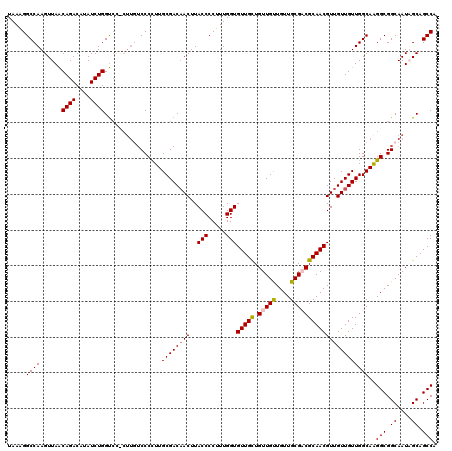
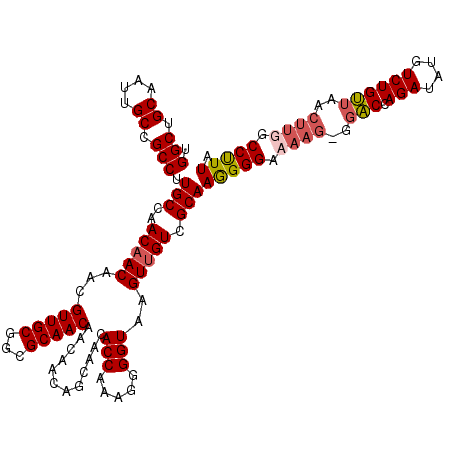
| Location | 21,280,475 – 21,280,594 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.93 |
| Mean single sequence MFE | -39.08 |
| Consensus MFE | -30.08 |
| Energy contribution | -31.08 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.750272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21280475 119 - 23771897 UGGCUGCUAUUGCCGCCCUGCCAACAACAACGUUGCGGCGCAACAACAACAACAACACCAAAGGGGUAAGUAGUCGCAAAGGGACAAG-GGACCAGAUAUGUCUGUUAACUUGGCCCUUA .(((((((..((((((.((((.(((......))))))).))................((...)))))))))))))...(((((.((((-..(.((((....)))).)..)))).))))). ( -38.40) >DroSec_CAF1 38742 120 - 1 UGGCUGCAAUUGCCGCCUUGCCAACAACAACGUUGCGUCGCAACAACAACAGCAACACCAAAGGGGUAAGUUGUCGCAAGGGGUAAAGGGGACCAGAUAUGUCUGUUAACUUGGCCUUUA .((((.....((((.((((((..(((((...(((((((.(......).)).)))))(((.....)))..))))).))))))))))(((..(((.(((....))))))..))))))).... ( -39.60) >DroSim_CAF1 40993 120 - 1 UGGCUGCAAUUGCCGCCUUGCCAACAACAACGUUGCGUCGCAACAACAACAGCAACACCAAAGGGGUAAGUUGUCGCAAGGGGACAAGGGGACCAGAUAUGUCUGUUAACUUGGCCUUUA .(((........((.((((((..(((((...(((((((.(......).)).)))))(((.....)))..))))).)))))))).((((..(((.(((....))))))..))))))).... ( -39.00) >DroEre_CAF1 39944 114 - 1 UGGCUGCUAUUGCCGCCUUGCCAACAACAACGUUGCGGCGCAACAGCAACAGCAACACCAAAGGGGUAAGUUGUCGCAAGGGGAA------GCCAGAUAUGUCUGCUAACUUGGCCUUUA .(((((((....((.((((((..(((((...(((((.((......))....)))))(((.....)))..))))).)))))))).)------))((((....)))).......)))).... ( -39.30) >consensus UGGCUGCAAUUGCCGCCUUGCCAACAACAACGUUGCGGCGCAACAACAACAGCAACACCAAAGGGGUAAGUUGUCGCAAGGGGAAAAG_GGACCAGAUAUGUCUGUUAACUUGGCCUUUA .(((.((....)).))).(((..(((((...(((((...)))))............(((.....)))..))))).)))(((((.((((..(((.(((....))))))..)))).))))). (-30.08 = -31.08 + 1.00)

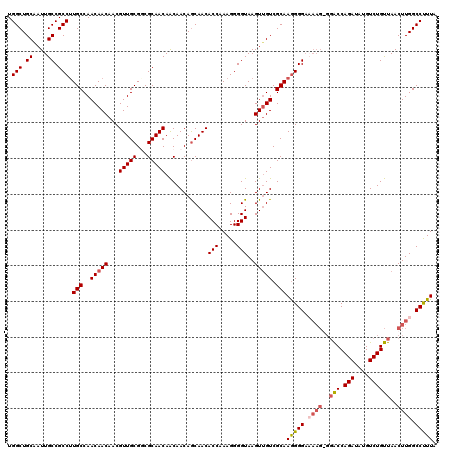
| Location | 21,280,514 – 21,280,634 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.79 |
| Mean single sequence MFE | -40.35 |
| Consensus MFE | -30.16 |
| Energy contribution | -30.97 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21280514 120 + 23771897 UUUGCGACUACUUACCCCUUUGGUGUUGUUGUUGUUGUUGCGCCGCAACGUUGUUGUUGGCAGGGCGGCAAUAGCAGCCAUUUCAAUAGUUGUGUUAAGUCAAUUGCAACUAGGUUUCUG .............(((...((((.((.((((((((((((((.((.(((((....)))))...)))))))))))))))).)).))))((((((((..........)))))))))))..... ( -35.90) >DroSec_CAF1 38782 116 + 1 CUUGCGACAACUUACCCCUUUGGUGUUGCUGUUGUUGUUGCGACGCAACGUUGUUGUUGGCAAGGCGGCAAUUGCAGCCAUUUCAAUAGUUGC----AGUCAAUUGCAACUAGGUUUCUG ((.((((((((..(((.....)))(((((.(((((....)))))))))))))))))).))...(((.((....)).))).......(((((((----((....)))))))))........ ( -40.70) >DroSim_CAF1 41033 117 + 1 CUUGCGACAACUUACCCCUUUGGUGUUGCUGUUGUUGUUGCGACGCAACGUUGUUGUUGGCAAGGCGGCAAUUGCAGCCAUUUCAAUAGUUGU---AAGUCAAUUGCAACUAGGUUUCUG ((.((((((((..(((.....)))(((((.(((((....)))))))))))))))))).))...(((.((....)).))).......(((((((---((.....)))))))))........ ( -39.80) >DroEre_CAF1 39978 120 + 1 CUUGCGACAACUUACCCCUUUGGUGUUGCUGUUGCUGUUGCGCCGCAACGUUGUUGUUGGCAAGGCGGCAAUAGCAGCCAUUUCAGCAGUAGUUGUUUGCGAAUUUCAACUAGGUUUCUG .(((((((((((.((..((..((((((((((((((((((..((((((((...)))).))))..)))))))))))))).))))..))..))))))).)))))).................. ( -45.00) >consensus CUUGCGACAACUUACCCCUUUGGUGUUGCUGUUGUUGUUGCGACGCAACGUUGUUGUUGGCAAGGCGGCAAUAGCAGCCAUUUCAAUAGUUGU___AAGUCAAUUGCAACUAGGUUUCUG ((.((((((((..(((.....)))(((((.(((((....)))))))))))))))))).))...(((.((....)).))).......(((((((............)))))))........ (-30.16 = -30.97 + 0.81)

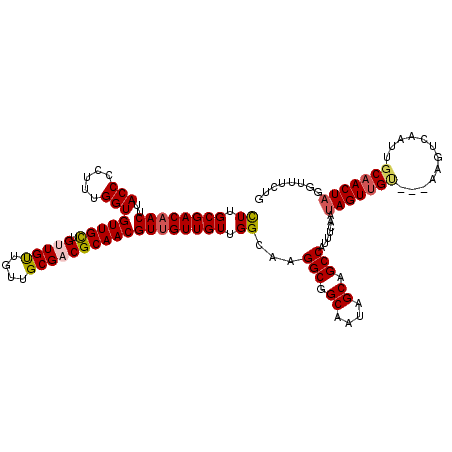
| Location | 21,280,514 – 21,280,634 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.79 |
| Mean single sequence MFE | -29.72 |
| Consensus MFE | -23.05 |
| Energy contribution | -23.61 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557414 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21280514 120 - 23771897 CAGAAACCUAGUUGCAAUUGACUUAACACAACUAUUGAAAUGGCUGCUAUUGCCGCCCUGCCAACAACAACGUUGCGGCGCAACAACAACAACAACACCAAAGGGGUAAGUAGUCGCAAA ...........((((..(((........)))..........(((((((..((((((.((((.(((......))))))).))................((...))))))))))))))))). ( -25.30) >DroSec_CAF1 38782 116 - 1 CAGAAACCUAGUUGCAAUUGACU----GCAACUAUUGAAAUGGCUGCAAUUGCCGCCUUGCCAACAACAACGUUGCGUCGCAACAACAACAGCAACACCAAAGGGGUAAGUUGUCGCAAG ........((((((((......)----))))))).......(((.((....)).)))((((..(((((...(((((((.(......).)).)))))(((.....)))..))))).)))). ( -32.80) >DroSim_CAF1 41033 117 - 1 CAGAAACCUAGUUGCAAUUGACUU---ACAACUAUUGAAAUGGCUGCAAUUGCCGCCUUGCCAACAACAACGUUGCGUCGCAACAACAACAGCAACACCAAAGGGGUAAGUUGUCGCAAG ...........((((...((((((---((..((.(((....(((.((....)).)))..............(((((((.(......).)).)))))..)))))..))))))))..)))). ( -28.00) >DroEre_CAF1 39978 120 - 1 CAGAAACCUAGUUGAAAUUCGCAAACAACUACUGCUGAAAUGGCUGCUAUUGCCGCCUUGCCAACAACAACGUUGCGGCGCAACAGCAACAGCAACACCAAAGGGGUAAGUUGUCGCAAG (((.....((((((...........))))))...)))....(((.((....)).)))((((..(((((...(((((.((......))....)))))(((.....)))..))))).)))). ( -32.80) >consensus CAGAAACCUAGUUGCAAUUGACUU___ACAACUAUUGAAAUGGCUGCAAUUGCCGCCUUGCCAACAACAACGUUGCGGCGCAACAACAACAGCAACACCAAAGGGGUAAGUUGUCGCAAG ........((((((..............)))))).......(((.((....)).)))((((..(((((...(((((...)))))............(((.....)))..))))).)))). (-23.05 = -23.61 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:06:09 2006