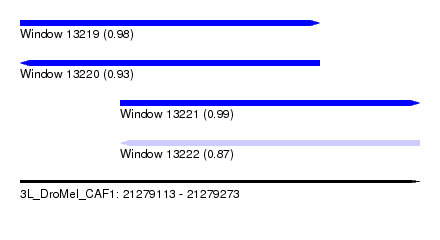
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,279,113 – 21,279,273 |
| Length | 160 |
| Max. P | 0.991827 |

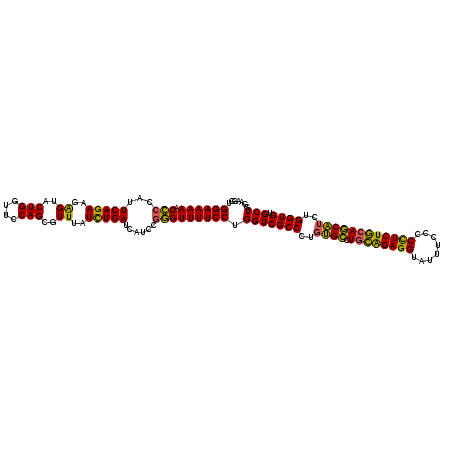
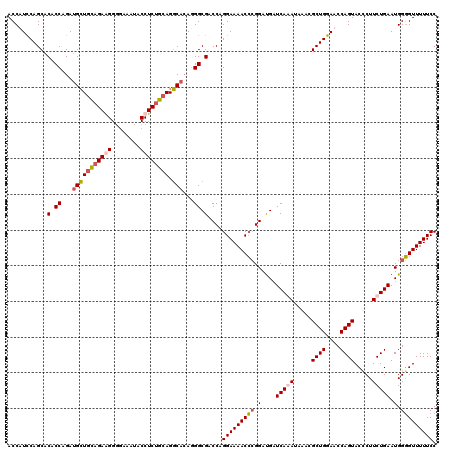
| Location | 21,279,113 – 21,279,233 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.58 |
| Mean single sequence MFE | -42.43 |
| Consensus MFE | -41.84 |
| Energy contribution | -40.15 |
| Covariance contribution | -1.69 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21279113 120 + 23771897 GGAAAAACCCCAUUCAGAAGAGUACUGGUUCCAGCGUUAAUCUGAUCAUCCGGGUUUUCCUGGUCGCCAUGUGCCUGCAGAGGUAUUUCCCCCUCUGCAGCAUCUGGUGUGCUGGAUGGU ((((((.(((...(((((..((..(((....)))..))..)))))......))))))))).((((((((.((((.((((((((........)))))))))))).))))).)))....... ( -49.00) >DroSec_CAF1 37355 120 + 1 GGAAAAACCCCAUUCAGAAGGGUACUGGUUCCAGCGUUUAUUUGAGCAUCCGGGUUUUCCUGGUCGCCCUUUGCCUGCAGAGGUAUUUCCCCUUCUGCAGCAUCUGGUGUGCUGAAAGGU ......(((.....((((((((..(((....)))...........((..(((((....)))))..))((((((....))))))......))))))))((((((.....))))))...))) ( -41.00) >DroSim_CAF1 39618 120 + 1 GGAAAAACCCCAUUCAGAAGGGUACUGGUUCCAGCGUUUAUUUGAUCGUCCGGGUUUUCCUGGUCGCCCUGUGCCUGCAGAGGUAUUUCCCCUUCUGCAGCAUCUGGUGUGCUGGAAGGU ......((((.........)))).....((((((((...........(.(((((....))))).)(((..((((.((((((((........))))))))))))..))).))))))))... ( -44.30) >DroEre_CAF1 38594 120 + 1 GGAAAAACACCAUUCAGAAGAGUACUGGUUCCAGCGUUUAUCUGAUCAUCCCUGUUUUCCUGGUCGCCUUGGGACUCUGGAGGUAUUUUGCCCUCUACAUCUUCUGGUGUGCUGGAUGGU ........(((((((((.((((((((...(((((.((((...((((((............)))))).....)))).)))))))))))))......((((((....))))))))))))))) ( -35.40) >consensus GGAAAAACCCCAUUCAGAAGAGUACUGGUUCCAGCGUUUAUCUGAUCAUCCGGGUUUUCCUGGUCGCCCUGUGCCUGCAGAGGUAUUUCCCCCUCUGCAGCAUCUGGUGUGCUGGAAGGU ((((((.(((...(((((..((..(((....)))..))..)))))......))))))))).(((((((..((((.((((((((........))))))))))))..)))).)))....... (-41.84 = -40.15 + -1.69)

| Location | 21,279,113 – 21,279,233 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.58 |
| Mean single sequence MFE | -39.68 |
| Consensus MFE | -31.52 |
| Energy contribution | -32.40 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933686 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21279113 120 - 23771897 ACCAUCCAGCACACCAGAUGCUGCAGAGGGGGAAAUACCUCUGCAGGCACAUGGCGACCAGGAAAACCCGGAUGAUCAGAUUAACGCUGGAACCAGUACUCUUCUGAAUGGGGUUUUUCC ...........(.(((..(((((((((((........)))))))).)))..))).)....(((((((((.(....(((((.....((((....)))).....))))).).))))))))). ( -45.60) >DroSec_CAF1 37355 120 - 1 ACCUUUCAGCACACCAGAUGCUGCAGAAGGGGAAAUACCUCUGCAGGCAAAGGGCGACCAGGAAAACCCGGAUGCUCAAAUAAACGCUGGAACCAGUACCCUUCUGAAUGGGGUUUUUCC .(((((((((((....).)))))..)))))(((((.(((((..((((....(((((.((.((....)).)).)))))........((((....)))).....))))...))))).))))) ( -41.40) >DroSim_CAF1 39618 120 - 1 ACCUUCCAGCACACCAGAUGCUGCAGAAGGGGAAAUACCUCUGCAGGCACAGGGCGACCAGGAAAACCCGGACGAUCAAAUAAACGCUGGAACCAGUACCCUUCUGAAUGGGGUUUUUCC .(((((((((((....).)))))..)))))(((((.(((((..((((...((((((.((.((....)).)).))...........((((....)))).))))))))...))))).))))) ( -41.90) >DroEre_CAF1 38594 120 - 1 ACCAUCCAGCACACCAGAAGAUGUAGAGGGCAAAAUACCUCCAGAGUCCCAAGGCGACCAGGAAAACAGGGAUGAUCAGAUAAACGCUGGAACCAGUACUCUUCUGAAUGGUGUUUUUCC ..........((((((.........((((........))))....(((((..((...)).(.....).)))))..(((((.....((((....)))).....))))).))))))...... ( -29.80) >consensus ACCAUCCAGCACACCAGAUGCUGCAGAAGGGGAAAUACCUCUGCAGGCACAGGGCGACCAGGAAAACCCGGAUGAUCAAAUAAACGCUGGAACCAGUACCCUUCUGAAUGGGGUUUUUCC ...........(.((...(((((((((((........)))))))).)))...)).)....(((((((((.(....(((((.....((((....)))).....))))).).))))))))). (-31.52 = -32.40 + 0.88)

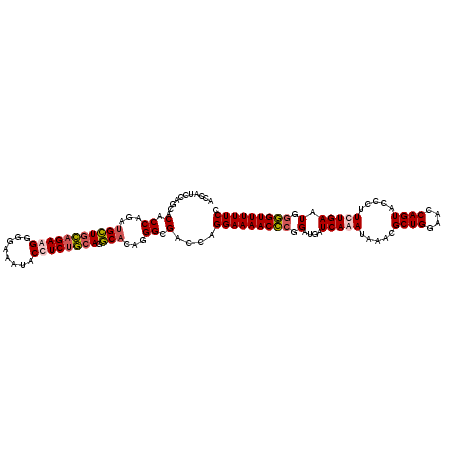
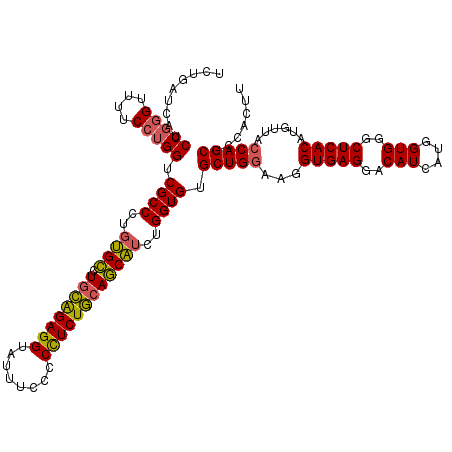
| Location | 21,279,153 – 21,279,273 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.14 |
| Mean single sequence MFE | -45.52 |
| Consensus MFE | -42.76 |
| Energy contribution | -42.45 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.991827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21279153 120 + 23771897 UCUGAUCAUCCGGGUUUUCCUGGUCGCCAUGUGCCUGCAGAGGUAUUUCCCCCUCUGCAGCAUCUGGUGUGCUGGAUGGUGAGGACAUCAUGGUGGGCUCACAUGUUACCAGCUCCACUU .........(((((....))))).(((((.((((.((((((((........)))))))))))).))))).(((((...(((((..(((....)))..)))))......)))))....... ( -51.70) >DroSec_CAF1 37395 120 + 1 UUUGAGCAUCCGGGUUUUCCUGGUCGCCCUUUGCCUGCAGAGGUAUUUCCCCUUCUGCAGCAUCUGGUGUGCUGAAAGGUGAGGACAUCAUGGUGGGCUCACAUGUUACCAGCUCCACUU ...((((..(((((....))))).((((...(((.((((((((........)))))))))))...))))((.(((...(((((..(((....)))..)))))...))).))))))..... ( -44.80) >DroSim_CAF1 39658 120 + 1 UUUGAUCGUCCGGGUUUUCCUGGUCGCCCUGUGCCUGCAGAGGUAUUUCCCCUUCUGCAGCAUCUGGUGUGCUGGAAGGUGAGGACAUCAUGGUGGGCUCACAUGUUACCAGCUCCACUU ..((((.((((..(..((((.(((((((..((((.((((((((........))))))))))))..)))).)))))))..)..)))))))).((((((((...........))).))))). ( -48.10) >DroEre_CAF1 38634 120 + 1 UCUGAUCAUCCCUGUUUUCCUGGUCGCCUUGGGACUCUGGAGGUAUUUUGCCCUCUACAUCUUCUGGUGUGCUGGAUGGUGAGGACAUCAUGGUGGGCUCACAGGUUACCAGCUCCACCU (((.(((((((..........(((((((..((((...((((((........))))))..))))..)))).)))))))))).))).......((((((((...........))).))))). ( -37.50) >consensus UCUGAUCAUCCGGGUUUUCCUGGUCGCCCUGUGCCUGCAGAGGUAUUUCCCCCUCUGCAGCAUCUGGUGUGCUGGAAGGUGAGGACAUCAUGGUGGGCUCACAUGUUACCAGCUCCACUU .........(((((....))))).((((..((((.((((((((........))))))))))))..)))).(((((...(((((..(((....)))..)))))......)))))....... (-42.76 = -42.45 + -0.31)

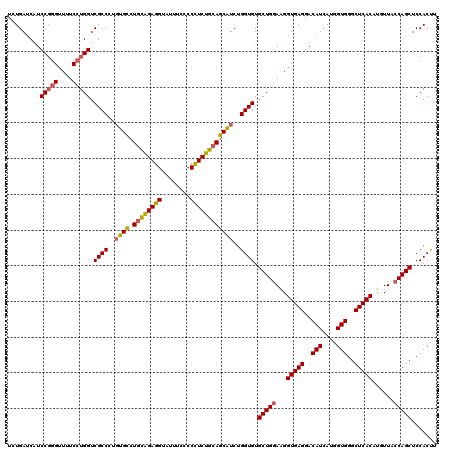
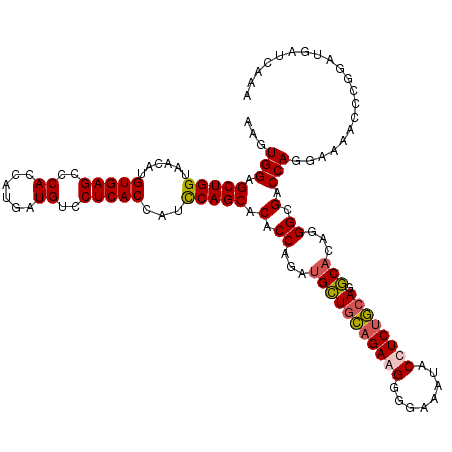
| Location | 21,279,153 – 21,279,273 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.14 |
| Mean single sequence MFE | -39.85 |
| Consensus MFE | -32.21 |
| Energy contribution | -32.52 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872434 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21279153 120 - 23771897 AAGUGGAGCUGGUAACAUGUGAGCCCACCAUGAUGUCCUCACCAUCCAGCACACCAGAUGCUGCAGAGGGGGAAAUACCUCUGCAGGCACAUGGCGACCAGGAAAACCCGGAUGAUCAGA ........(((((.....(((((..((......))..)))))(((((....(.(((..(((((((((((........)))))))).)))..))).)....((.....)))))))))))). ( -43.80) >DroSec_CAF1 37395 120 - 1 AAGUGGAGCUGGUAACAUGUGAGCCCACCAUGAUGUCCUCACCUUUCAGCACACCAGAUGCUGCAGAAGGGGAAAUACCUCUGCAGGCAAAGGGCGACCAGGAAAACCCGGAUGCUCAAA ..(((..(((((......(((((..((......))..)))))...))))).)))....(((((((((.((.......)))))))).)))..(((((.((.((....)).)).)))))... ( -40.80) >DroSim_CAF1 39658 120 - 1 AAGUGGAGCUGGUAACAUGUGAGCCCACCAUGAUGUCCUCACCUUCCAGCACACCAGAUGCUGCAGAAGGGGAAAUACCUCUGCAGGCACAGGGCGACCAGGAAAACCCGGACGAUCAAA ..((((.(((.(.......).)))))))..((((((((.....((((...........(((((((((.((.......)))))))).)))..((....)).)))).....)))).)))).. ( -39.40) >DroEre_CAF1 38634 120 - 1 AGGUGGAGCUGGUAACCUGUGAGCCCACCAUGAUGUCCUCACCAUCCAGCACACCAGAAGAUGUAGAGGGCAAAAUACCUCCAGAGUCCCAAGGCGACCAGGAAAACAGGGAUGAUCAGA .((((..(((((......(((((..((......))..)))))...))))).))))....(((...((((........))))....(((((..((...)).(.....).))))).)))... ( -35.40) >consensus AAGUGGAGCUGGUAACAUGUGAGCCCACCAUGAUGUCCUCACCAUCCAGCACACCAGAUGCUGCAGAAGGGGAAAUACCUCUGCAGGCACAGGGCGACCAGGAAAACCCGGAUGAUCAAA ...(((.(((((......(((((..((......))..)))))...))))).(.((...(((((((((((........)))))))).)))...)).).))).................... (-32.21 = -32.52 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:06:00 2006