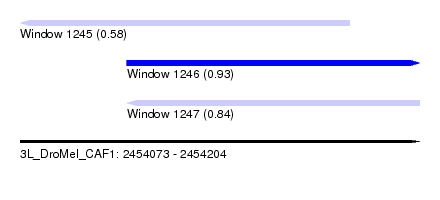
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,454,073 – 2,454,204 |
| Length | 131 |
| Max. P | 0.933632 |

| Location | 2,454,073 – 2,454,181 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.99 |
| Mean single sequence MFE | -27.22 |
| Consensus MFE | -19.81 |
| Energy contribution | -21.01 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
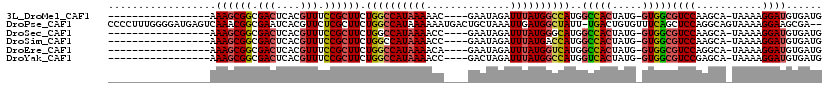
>3L_DroMel_CAF1 2454073 108 - 23771897 UUCUGGCCAUAAAAAC----GAAUAGAUUUAUGGCCAUGGCCACUAUG-GUGGCGUCCAAGCA-UAAAAGGAUGUGAUGCGCUGGCCGAAA-AAUGACUCAUG---AAAUGUGCA-CUA (((((((((((((...----.......)))))))))).(((((.....-((.((((((.....-.....)))))).))....)))))))).-...........---.........-... ( -30.20) >DroPse_CAF1 87310 103 - 1 UUCUGGCCAUAAAAAAUGACUGCUAAAUUGAUGGCUAUU-UGACUGUGUUUCAGCUCCAGGCAGUAAAAGGAAGCGA----AUGGCCAAAACUAUGACUCACCAUCAA----------- ...(((((((........((((((....((..((((...-.(((...)))..)))).))))))))....(....)..----)))))))....................----------- ( -19.50) >DroSec_CAF1 77779 108 - 1 UUCUGGCCAUAAAACC----GAAUAGAUUUAUGGGCAUGGCCACUAUG-GUGGCGUCCAAGCA-UAAAAGGAUGUGAUGCGCUGGCCGAAA-AAUGACUCAUG---AAAUGUGCA-CUA ......(((((((.(.----.....).)))))))(((((((((.....-((.((((((.....-.....)))))).))....)))))((..-......))...---....)))).-... ( -26.60) >DroSim_CAF1 71812 109 - 1 UUCUGGCCAUAAAACC----GAAUAGAUUUAUGACCAUGGCCACUAUG-GUGGCGUCCAAGCA-UAAAAGGAUGUGAUGCGCUGGCCGAAA-AAUGACUCAUG---AAAUGUGCAACUA (((.((((........----.............(((((((...)))))-))(((((....(((-(......))))...)))))))))))).-...........---............. ( -24.70) >DroEre_CAF1 72032 107 - 1 UUCUGGCCAUAAAACA----GAAUAGAUUUAUGGUCAUGGCCACUAUG-GUGGCGUCCAGGCA-UAAAAGGAUGUGAUGCGCUGGCC-AAA-AAUGACUCAUG---AAAUAUGCA-CUA (((((.........))----)))....(((((((((((.(((((....-)))))(.(((((((-(...........)))).)))).)-...-.))))).))))---)).......-... ( -31.90) >DroYak_CAF1 74966 96 - 1 UUCUGGCCAUAAAACC----GACUAGAUUUAUGGCCAUGGUCACUAUG-GUGGCGUCCGAGCA-UAAAAGGAUGUGAUGCGCUGGCC-AAA-AAUGACUCAUG---A------------ ....(((((((((.(.----.....).)))))))))(((((((...((-(((((((....(((-(......))))...))))).)))-)..-..)))).))).---.------------ ( -30.40) >consensus UUCUGGCCAUAAAACC____GAAUAGAUUUAUGGCCAUGGCCACUAUG_GUGGCGUCCAAGCA_UAAAAGGAUGUGAUGCGCUGGCCGAAA_AAUGACUCAUG___AAAUGUGCA_CUA ...((((((((((..............)))))))))).(((((......((.((((((...........)))))).))....)))))................................ (-19.81 = -21.01 + 1.20)

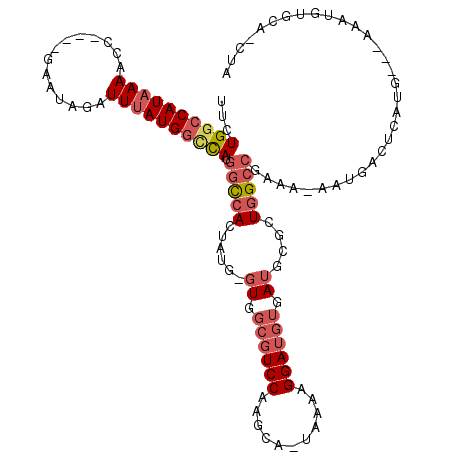
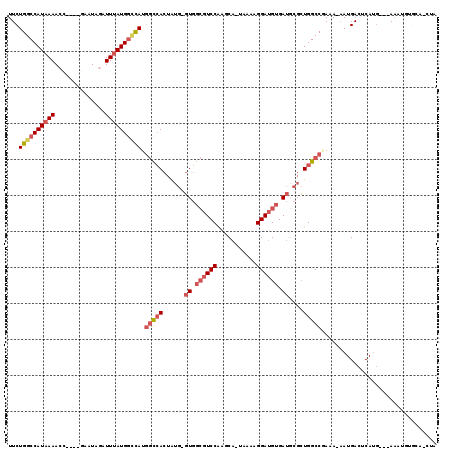
| Location | 2,454,108 – 2,454,204 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.41 |
| Mean single sequence MFE | -32.63 |
| Consensus MFE | -23.00 |
| Energy contribution | -26.39 |
| Covariance contribution | 3.39 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933632 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2454108 96 + 23771897 CAUCACAUCCUUUUA-UGCUUGGACGCCAC-CAUAGUGGCCAUGGCCAUAAAUCUAUUC----GUUUUUAUGGCCAGAAGCGGAAACGUGAGUCGCCGCUUU----------------- .......(((.....-.....))).(((((-....)))))..((((((((((.......----...))))))))))(((((((..((....))..)))))))----------------- ( -37.30) >DroPse_CAF1 87337 116 + 1 --UCGCUUCCUUUUACUGCCUGGAGCUGAAACACAGUCA-AAUAGCCAUCAAUUUAGCAGUCAUUUUUUAUGGCCAGAAGCGAGAACGUGAUUCGCCGUUUGACUCAUCCCCAAAGGGG --..(((.((...........)))))........(((((-((..((.((((.(((.((.(((((.....))))).....)).)))...))))..))..)))))))...((((...)))) ( -25.50) >DroSec_CAF1 77814 96 + 1 CAUCACAUCCUUUUA-UGCUUGGACGCCAC-CAUAGUGGCCAUGCCCAUAAAUCUAUUC----GGUUUUAUGGCCAGAAGCGGAAACGUGAGUCGCCGCUUU----------------- .......(((.....-.....))).(((((-....)))))..((.(((((((.((....----)).))))))).))(((((((..((....))..)))))))----------------- ( -31.40) >DroSim_CAF1 71848 96 + 1 CAUCACAUCCUUUUA-UGCUUGGACGCCAC-CAUAGUGGCCAUGGUCAUAAAUCUAUUC----GGUUUUAUGGCCAGAAGCGGAAACGUGAGUCGCCGCUUU----------------- .......(((.....-.....))).(((((-....)))))..((((((((((.((....----)).))))))))))(((((((..((....))..)))))))----------------- ( -35.80) >DroEre_CAF1 72066 96 + 1 CAUCACAUCCUUUUA-UGCCUGGACGCCAC-CAUAGUGGCCAUGACCAUAAAUCUAUUC----UGUUUUAUGGCCAGAAGCGGAAACGUGAGUCGCCGCUUU----------------- ...............-...((((..(((((-....))))).....(((((((.(.....----.).)))))))))))((((((..((....))..)))))).----------------- ( -32.80) >DroYak_CAF1 74989 96 + 1 CAUCACAUCCUUUUA-UGCUCGGACGCCAC-CAUAGUGACCAUGGCCAUAAAUCUAGUC----GGUUUUAUGGCCAGAAGCGGAAACGUGAGUCGCCGCUUU----------------- ...............-.....((.(((...-....))).)).((((((((((.((....----)).))))))))))(((((((..((....))..)))))))----------------- ( -33.00) >consensus CAUCACAUCCUUUUA_UGCUUGGACGCCAC_CAUAGUGGCCAUGGCCAUAAAUCUAUUC____GGUUUUAUGGCCAGAAGCGGAAACGUGAGUCGCCGCUUU_________________ .......(((...........))).(((((.....)))))..((((((((((..............))))))))))(((((((..((....))..)))))))................. (-23.00 = -26.39 + 3.39)

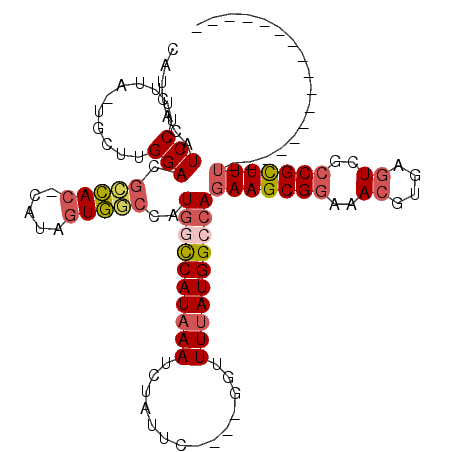
| Location | 2,454,108 – 2,454,204 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.41 |
| Mean single sequence MFE | -31.23 |
| Consensus MFE | -24.75 |
| Energy contribution | -25.06 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
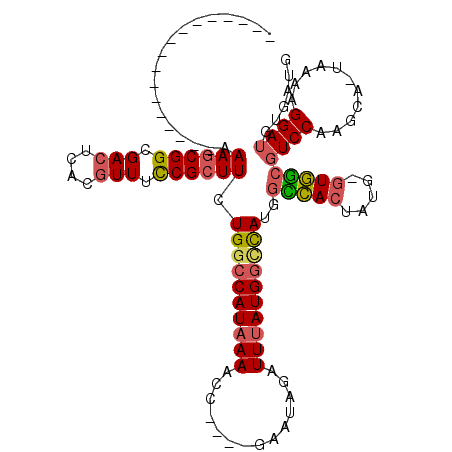
>3L_DroMel_CAF1 2454108 96 - 23771897 -----------------AAAGCGGCGACUCACGUUUCCGCUUCUGGCCAUAAAAAC----GAAUAGAUUUAUGGCCAUGGCCACUAUG-GUGGCGUCCAAGCA-UAAAAGGAUGUGAUG -----------------.((((((.(((....))).)))))).((((((((((...----.......))))))))))..(((((....-)))))((((.....-.....))))...... ( -34.10) >DroPse_CAF1 87337 116 - 1 CCCCUUUGGGGAUGAGUCAAACGGCGAAUCACGUUCUCGCUUCUGGCCAUAAAAAAUGACUGCUAAAUUGAUGGCUAUU-UGACUGUGUUUCAGCUCCAGGCAGUAAAAGGAAGCGA-- (((....)))(((..(((....)))..)))......((((((((((((((....(((.........))).)))))).((-(.(((((.(.........).))))).)))))))))))-- ( -30.00) >DroSec_CAF1 77814 96 - 1 -----------------AAAGCGGCGACUCACGUUUCCGCUUCUGGCCAUAAAACC----GAAUAGAUUUAUGGGCAUGGCCACUAUG-GUGGCGUCCAAGCA-UAAAAGGAUGUGAUG -----------------.((((((.(((....))).)))))).((.(((((((.(.----.....).))))))).))..(((((....-)))))((((.....-.....))))...... ( -30.20) >DroSim_CAF1 71848 96 - 1 -----------------AAAGCGGCGACUCACGUUUCCGCUUCUGGCCAUAAAACC----GAAUAGAUUUAUGACCAUGGCCACUAUG-GUGGCGUCCAAGCA-UAAAAGGAUGUGAUG -----------------.((((((.(((....))).)))))).(((.((((((.(.----.....).)))))).)))..(((((....-)))))((((.....-.....))))...... ( -27.70) >DroEre_CAF1 72066 96 - 1 -----------------AAAGCGGCGACUCACGUUUCCGCUUCUGGCCAUAAAACA----GAAUAGAUUUAUGGUCAUGGCCACUAUG-GUGGCGUCCAGGCA-UAAAAGGAUGUGAUG -----------------.((((((.(((....))).)))))).((((((((((.(.----.....).))))))))))..(((((....-)))))((((.....-.....))))...... ( -32.60) >DroYak_CAF1 74989 96 - 1 -----------------AAAGCGGCGACUCACGUUUCCGCUUCUGGCCAUAAAACC----GACUAGAUUUAUGGCCAUGGUCACUAUG-GUGGCGUCCGAGCA-UAAAAGGAUGUGAUG -----------------.((((((.(((....))).)))))).((((((((((.(.----.....).))))))))))..(((((....-)))))((((.....-.....))))...... ( -32.80) >consensus _________________AAAGCGGCGACUCACGUUUCCGCUUCUGGCCAUAAAACC____GAAUAGAUUUAUGGCCAUGGCCACUAUG_GUGGCGUCCAAGCA_UAAAAGGAUGUGAUG ..................((((((.(((....))).)))))).((((((((((..............))))))))))..(((((.....)))))((((...........))))...... (-24.75 = -25.06 + 0.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:45 2006