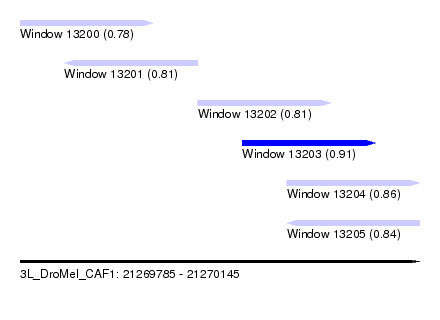
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,269,785 – 21,270,145 |
| Length | 360 |
| Max. P | 0.906951 |

| Location | 21,269,785 – 21,269,905 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -42.16 |
| Consensus MFE | -40.01 |
| Energy contribution | -39.38 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.782852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21269785 120 + 23771897 CAGGGAAUACCCACUCCGGUAGUAGCCCUCAUGGAUCUGGAGUCAAUCAAUCCCAAGCGUCGUCCUGCUGCCAAGCAGGCGGCUCAGAAGGUCAUAUCUAAGCUGUUGCCCGAGGAGAAG ..(((....))).(((((((((((((.......((((((((.........)))...(.((((.((((((....)))))))))).)...)))))........)))))))))...))))... ( -41.66) >DroSec_CAF1 28062 120 + 1 CAGGGAAUACCCACUCCGGUGGUAGCCCUCAUGGAUCUGGAGUCAAUCAAUCCCAAGCGGCGUCCUGCUACCAAGCAGGCGGCUCAGAAGAUCAUAUCCAAGCUGUUACCCGAGGAGAAG ..(((....))).(((((((..((((.......(((((.(((((........((....)).(.((((((....))))))))))))...)))))........))))..)))...))))... ( -43.16) >DroSim_CAF1 30318 120 + 1 CAGGGAAUACCCACUCCGGUGGUAGCCCUCAUGGAUCUGGAGUCAAUCAAUCCCAAGCGGCGUCCUGCUACCAAGCAGGCGGCUCAGAAGAUCAUAUCCAAGCUGUUACCCGAGGAGAAG ..(((....))).(((((((..((((.......(((((.(((((........((....)).(.((((((....))))))))))))...)))))........))))..)))...))))... ( -43.16) >DroEre_CAF1 29237 120 + 1 CAGGGAAUACCCACUCCGGUGGUAGCCCUCAUGGAUCUGGAGUCAAUCAAUCCCAAGCGUCGUUCUGCUGCCAAGCAGGCGGCUCAAAAGAUCAUCUCAAAGCUGUUGCCCGAGGAGAAG ..(((....))).(((((((..((((.......(((((.(((((...........((((......))))(((.....))))))))...)))))........))))..)))...))))... ( -40.66) >consensus CAGGGAAUACCCACUCCGGUGGUAGCCCUCAUGGAUCUGGAGUCAAUCAAUCCCAAGCGGCGUCCUGCUACCAAGCAGGCGGCUCAGAAGAUCAUAUCCAAGCUGUUACCCGAGGAGAAG ..(((....))).(((((((..((((.......(((((.(((((.........(....)..(.((((((....))))))))))))...)))))........))))..)))...))))... (-40.01 = -39.38 + -0.62)


| Location | 21,269,825 – 21,269,945 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.75 |
| Mean single sequence MFE | -40.45 |
| Consensus MFE | -37.38 |
| Energy contribution | -37.38 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812404 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21269825 120 - 23771897 UAACAUUCAAGGCCUCGCUAGCGGUGUCCAAUUGCAUGAUCUUCUCCUCGGGCAACAGCUUAGAUAUGACCUUCUGAGCCGCCUGCUUGGCAGCAGGACGACGCUUGGGAUUGAUUGACU ...((.((((..(((....((((.(((((..((((..((.....))..((((((.(.(((((((........))))))).)..))))))))))..))))).)))).))).)))).))... ( -39.30) >DroSec_CAF1 28102 120 - 1 UCACAUUCAAGCCUUCGCUAGCGGUGUCCAAUUGCAUGAUCUUCUCCUCGGGUAACAGCUUGGAUAUGAUCUUCUGAGCCGCCUGCUUGGUAGCAGGACGCCGCUUGGGAUUGAUUGACU ...((.((((.(((.....((((((((((........((((...(((...(((....))).)))...))))......((..((.....))..)).)))))))))).))).)))).))... ( -43.00) >DroSim_CAF1 30358 120 - 1 UCACAUUCAAGCCUUCGCUAGCGGUGUCCAAUUGCAUGAUCUUCUCCUCGGGUAACAGCUUGGAUAUGAUCUUCUGAGCCGCCUGCUUGGUAGCAGGACGCCGCUUGGGAUUGAUUGACU ...((.((((.(((.....((((((((((........((((...(((...(((....))).)))...))))......((..((.....))..)).)))))))))).))).)))).))... ( -43.00) >DroEre_CAF1 29277 120 - 1 UCACAUUUAAACCCUCGCUAGCGGUGUCCAAUUGCAUGAUCUUCUCCUCGGGCAACAGCUUUGAGAUGAUCUUUUGAGCCGCCUGCUUGGCAGCAGAACGACGCUUGGGAUUGAUUGACU ...((.((((.(((..((..(((((...(((......((((.((((...((((....)))).)))).))))..))).)))))(((((....)))))......))..))).)))).))... ( -36.50) >consensus UCACAUUCAAGCCCUCGCUAGCGGUGUCCAAUUGCAUGAUCUUCUCCUCGGGCAACAGCUUGGAUAUGAUCUUCUGAGCCGCCUGCUUGGCAGCAGGACGACGCUUGGGAUUGAUUGACU ...((.((((.(((.....((((((((((..((((..((.....))..((((((.(.(((((((........))))))).)..))))))))))..)))))))))).))).)))).))... (-37.38 = -37.38 + 0.00)


| Location | 21,269,945 – 21,270,065 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.61 |
| Mean single sequence MFE | -43.44 |
| Consensus MFE | -38.67 |
| Energy contribution | -38.11 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21269945 120 + 23771897 UGCGGCGCAUUGGAGGCCAGAAGAAGCGCAUCCUCCAUAACGUCGCCAAUCGGCCACAUCUCUUUGGAGAUGUGGUAGAGUUUAAACCGGGCUCAGAUCCCAGCGACGAUCUGGGCACAC .((((((...((((((...(......)....))))))...))))))...((.((((((((((....)))))))))).))...........(((((((((........))))))))).... ( -50.90) >DroSec_CAF1 28222 120 + 1 UGCGACGCAUUGGCGGCCAGAAGAAGCGCAUCCUCCAUAACGUCGCCAAUCGGCCACAUCUCUUUGGCGAUGUGGUAGAGUUCAAACUGGGUUCAGAUUCCAGCGACGAUCUGGGCACCC (((..(((....))).(((((.((......))........((((((.((((.((((((((.(....).)))))))).(((..(.....)..))).))))...)))))).))))))))... ( -41.10) >DroSim_CAF1 30478 120 + 1 UGCGACGCAUUGGUGGCCAGAAGAAGCGCAUCCUCCAUAACGUCGCCAAUCGGCCACAUCUCUUUGGCGAUGUGGUAGAGUUCAAACCGGGUUCAGAUCCCAGCGACGAUCUGGGCACCC ...........((((.(((((.....(((...(((.((.((((((((((..((......))..)))))))))).)).)))........(((.......))).)))....))))).)))). ( -44.20) >DroEre_CAF1 29397 120 + 1 UGCGACGAAUUGGCGGCCAAAAGAAGCGCAUCCUCCAUAACGUCGCCAAUCGGCCACAUCUCUUUGGCGAUGUGGUAGAGUUUAAACCAAGCUCAAAUCCCAGCGACGAUCUGGGUACCC .((((((...(((.((...............)).)))...))))))......((((((((.(....).)))))))).((((((.....))))))....(((((.......)))))..... ( -37.56) >consensus UGCGACGCAUUGGCGGCCAGAAGAAGCGCAUCCUCCAUAACGUCGCCAAUCGGCCACAUCUCUUUGGCGAUGUGGUAGAGUUCAAACCGGGCUCAGAUCCCAGCGACGAUCUGGGCACCC .((((((...(((.((...............)).)))...))))))...((.((((((((.(....).)))))))).))...........(((((((((........))))))))).... (-38.67 = -38.11 + -0.56)



| Location | 21,269,985 – 21,270,105 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -41.38 |
| Consensus MFE | -36.65 |
| Energy contribution | -35.65 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21269985 120 + 23771897 CGUCGCCAAUCGGCCACAUCUCUUUGGAGAUGUGGUAGAGUUUAAACCGGGCUCAGAUCCCAGCGACGAUCUGGGCACACUAGAAGUUACUGGUUUCUUACGUGGACAGUCUCUUAACGU .(((.....((.((((((((((....)))))))))).))((..((((((((((((((((........)))))))))..((.....))..)))))))...))...)))............. ( -43.40) >DroSec_CAF1 28262 120 + 1 CGUCGCCAAUCGGCCACAUCUCUUUGGCGAUGUGGUAGAGUUCAAACUGGGUUCAGAUUCCAGCGACGAUCUGGGCACCCUAGAAGUUACUGGUUUCUUACGUGGACAGUCUCUAAACGU (((((((((..((......))..)))))))))...(((((......(((.((..(((..((((..((..((((((...)))))).))..))))..))).)).))).....)))))..... ( -39.70) >DroSim_CAF1 30518 120 + 1 CGUCGCCAAUCGGCCACAUCUCUUUGGCGAUGUGGUAGAGUUCAAACCGGGUUCAGAUCCCAGCGACGAUCUGGGCACCCUAGAAGUUACUGGUUUCUUACGUGGACAGUCUCUAAACGU (((((((((..((......))..)))))))))...(((((......(((.((..(((..((((..((..((((((...)))))).))..))))..))).)).))).....)))))..... ( -42.80) >DroEre_CAF1 29437 120 + 1 CGUCGCCAAUCGGCCACAUCUCUUUGGCGAUGUGGUAGAGUUUAAACCAAGCUCAAAUCCCAGCGACGAUCUGGGUACCCUAGAAGUCACUGGAUUCUUACGCGGACAGUCUCUAAACGU .(((((......((((((((.(....).)))))))).((((((.....)))))).....((((.(((..((((((...)))))).))).))))........)).)))............. ( -39.60) >consensus CGUCGCCAAUCGGCCACAUCUCUUUGGCGAUGUGGUAGAGUUCAAACCGGGCUCAGAUCCCAGCGACGAUCUGGGCACCCUAGAAGUUACUGGUUUCUUACGUGGACAGUCUCUAAACGU .(((((......((((((((.(....).)))))))).((((((.....))))))(((..((((.(((..(((((.....))))).))).))))..)))...)).)))............. (-36.65 = -35.65 + -1.00)


| Location | 21,270,025 – 21,270,145 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.47 |
| Mean single sequence MFE | -40.83 |
| Consensus MFE | -34.16 |
| Energy contribution | -34.85 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21270025 120 + 23771897 UUUAAACCGGGCUCAGAUCCCAGCGACGAUCUGGGCACACUAGAAGUUACUGGUUUCUUACGUGGACAGUCUCUUAACGUGAAUGGUUUGGUCCACAUUCCCGGCUUGGGUGAUUUCCAG ......(((((...(((..((((..((..(((((.....))))).))..))))..)))...((((((...((.((......)).))....))))))...)))))..((((.....)))). ( -39.60) >DroSec_CAF1 28302 120 + 1 UUCAAACUGGGUUCAGAUUCCAGCGACGAUCUGGGCACCCUAGAAGUUACUGGUUUCUUACGUGGACAGUCUCUAAACGUGAAUGGUUUGGUCCACAUUCCCGGUUUGGGUGAUUUCCAG ..(((((((((...(((..((((..((..((((((...)))))).))..))))..)))...((((((.....(((........)))....))))))...)))))))))((......)).. ( -44.10) >DroSim_CAF1 30558 120 + 1 UUCAAACCGGGUUCAGAUCCCAGCGACGAUCUGGGCACCCUAGAAGUUACUGGUUUCUUACGUGGACAGUCUCUAAACGUGAAUGGUUUGGUCCACAUUCCCGGCUUGGGUGAUUUCCAG ..(((.(((((...(((..((((..((..((((((...)))))).))..))))..)))...((((((.....(((........)))....))))))...))))).)))((......)).. ( -42.30) >DroEre_CAF1 29477 120 + 1 UUUAAACCAAGCUCAAAUCCCAGCGACGAUCUGGGUACCCUAGAAGUCACUGGAUUCUUACGCGGACAGUCUCUAAACGUGAAUGGUUUGGUUCACAUUCCCGGCUUGGGUGAUUUCCAG ......((((((.......((((.(((..((((((...)))))).))).)))).......((.(((..((..((((((.......))))))...))..)))))))))))........... ( -37.30) >consensus UUCAAACCGGGCUCAGAUCCCAGCGACGAUCUGGGCACCCUAGAAGUUACUGGUUUCUUACGUGGACAGUCUCUAAACGUGAAUGGUUUGGUCCACAUUCCCGGCUUGGGUGAUUUCCAG ......(((((...(((..((((.(((..(((((.....))))).))).))))..)))...((((((.....(((........)))....))))))...)))))..((((.....)))). (-34.16 = -34.85 + 0.69)



| Location | 21,270,025 – 21,270,145 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.47 |
| Mean single sequence MFE | -37.45 |
| Consensus MFE | -32.66 |
| Energy contribution | -32.48 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21270025 120 - 23771897 CUGGAAAUCACCCAAGCCGGGAAUGUGGACCAAACCAUUCACGUUAAGAGACUGUCCACGUAAGAAACCAGUAACUUCUAGUGUGCCCAGAUCGUCGCUGGGAUCUGAGCCCGGUUUAAA ..((.......))((((((((.((((((((.......................)))))))).(((..((((..((.(((.(.....).)))..))..))))..)))...))))))))... ( -35.40) >DroSec_CAF1 28302 120 - 1 CUGGAAAUCACCCAAACCGGGAAUGUGGACCAAACCAUUCACGUUUAGAGACUGUCCACGUAAGAAACCAGUAACUUCUAGGGUGCCCAGAUCGUCGCUGGAAUCUGAACCCAGUUUGAA ............(((((.(((.((((((((.......................)))))))).(((..((((..((.(((.((....)))))..))..))))..)))...))).))))).. ( -34.60) >DroSim_CAF1 30558 120 - 1 CUGGAAAUCACCCAAGCCGGGAAUGUGGACCAAACCAUUCACGUUUAGAGACUGUCCACGUAAGAAACCAGUAACUUCUAGGGUGCCCAGAUCGUCGCUGGGAUCUGAACCCGGUUUGAA ............(((((((((.((((((((.......................)))))))).(((..((((..((.(((.((....)))))..))..))))..)))...))))))))).. ( -42.70) >DroEre_CAF1 29477 120 - 1 CUGGAAAUCACCCAAGCCGGGAAUGUGAACCAAACCAUUCACGUUUAGAGACUGUCCGCGUAAGAAUCCAGUGACUUCUAGGGUACCCAGAUCGUCGCUGGGAUUUGAGCUUGGUUUAAA ..(((..((.(((.....)))((((((((........))))))))....))...)))((...(((.(((((((((.(((.((....)))))..))))))))).)))..)).......... ( -37.10) >consensus CUGGAAAUCACCCAAGCCGGGAAUGUGGACCAAACCAUUCACGUUUAGAGACUGUCCACGUAAGAAACCAGUAACUUCUAGGGUGCCCAGAUCGUCGCUGGGAUCUGAACCCGGUUUAAA ..((.......))((((((((.((((((((.......................)))))))).(((..(((((.((.(((.((....)))))..)).)))))..)))...))))))))... (-32.66 = -32.48 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:05:43 2006