
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,269,145 – 21,269,345 |
| Length | 200 |
| Max. P | 0.939537 |

| Location | 21,269,145 – 21,269,265 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.14 |
| Mean single sequence MFE | -34.67 |
| Consensus MFE | -30.00 |
| Energy contribution | -29.62 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.541780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21269145 120 + 23771897 GUUAAUUACCUGGUAACUAUGGCUGAUCAUGCUUUUCAUCGGCCCGGGCCGCUGAAACAAGCGAACAAGGCUCACAAAACGGGUCGUCAUCGCUCCAAAGGAGCAAUAGACAAUGCCCAA ....................(((((((.(......).))))))).((((((((......)))).....(((((.......)))))(((((.(((((...))))).)).)))...)))).. ( -37.10) >DroSec_CAF1 27422 120 + 1 GUUAAUUACCUGGUAACUAUGGCGGAUCAUGCAUUUCAUCGGCCCGGGCCGCUGAAACAAACGAACAAGGCUCACAAAACAGGUCGUCAUCGCUCCAAAGGAGCAAUAGACAAUGCCCAA .......(((((........(((.(((.(......).))).))).(((((..((...........)).)))))......))))).(((((.(((((...))))).)).)))......... ( -31.00) >DroSim_CAF1 29678 120 + 1 GUUAAUUACCUGCUAACUAUGGCGGAUCAUGCAUUUCAUCGGCCCGGGCCGCUGAAACAAACGAACAAGGCUCACAAAACAGGUCGUCAUCGCUCCAAAGGGGCAAUAGACAAUGCCCAA .........((((((....)))))).....((((((((.((((....)))).)))))...........((((.........))))(((((.(((((...))))).)).)))..))).... ( -31.00) >DroEre_CAF1 28598 119 + 1 GUUAAAUACCUG-UGACAAUGGCGGAUCAUGCGUUUCAUCGGCCCGGGCCGCUGAAGCAAACGAACAAGGCUCACAAAACGGGUCGUCAUCGCUCCAAAGGAGCAAUAGACAAUGCCCAA ..........((-((((..(((((.....)))((((((.((((....)))).)))))).......))..).)))))....((((.(((((.(((((...))))).)).)))...)))).. ( -39.60) >consensus GUUAAUUACCUGGUAACUAUGGCGGAUCAUGCAUUUCAUCGGCCCGGGCCGCUGAAACAAACGAACAAGGCUCACAAAACAGGUCGUCAUCGCUCCAAAGGAGCAAUAGACAAUGCCCAA ....................(((.((((.....(((((.((((....)))).))))).....((.......))........))))(((((.(((((...))))).)).)))...)))... (-30.00 = -29.62 + -0.38)

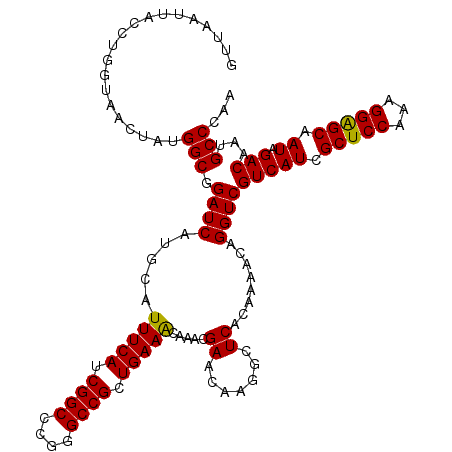

| Location | 21,269,145 – 21,269,265 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.14 |
| Mean single sequence MFE | -41.12 |
| Consensus MFE | -36.67 |
| Energy contribution | -37.55 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21269145 120 - 23771897 UUGGGCAUUGUCUAUUGCUCCUUUGGAGCGAUGACGACCCGUUUUGUGAGCCUUGUUCGCUUGUUUCAGCGGCCCGGGCCGAUGAAAAGCAUGAUCAGCCAUAGUUACCAGGUAAUUAAC ...(((.(((((.((((((((...)))))))))))))...(((((((((((...)))))).....(((.((((....)))).)))))))).......))).(((((((...))))))).. ( -41.50) >DroSec_CAF1 27422 120 - 1 UUGGGCAUUGUCUAUUGCUCCUUUGGAGCGAUGACGACCUGUUUUGUGAGCCUUGUUCGUUUGUUUCAGCGGCCCGGGCCGAUGAAAUGCAUGAUCCGCCAUAGUUACCAGGUAAUUAAC ........((((.((((((((...))))))))))))(((((..(((((.((.....((((.(((((((.((((....)))).))))))).))))...)))))))....)))))....... ( -42.30) >DroSim_CAF1 29678 120 - 1 UUGGGCAUUGUCUAUUGCCCCUUUGGAGCGAUGACGACCUGUUUUGUGAGCCUUGUUCGUUUGUUUCAGCGGCCCGGGCCGAUGAAAUGCAUGAUCCGCCAUAGUUAGCAGGUAAUUAAC ........((((.(((((((....)).)))))))))((((((((((((.((.....((((.(((((((.((((....)))).))))))).))))...)))))))..)))))))....... ( -40.50) >DroEre_CAF1 28598 119 - 1 UUGGGCAUUGUCUAUUGCUCCUUUGGAGCGAUGACGACCCGUUUUGUGAGCCUUGUUCGUUUGCUUCAGCGGCCCGGGCCGAUGAAACGCAUGAUCCGCCAUUGUCA-CAGGUAUUUAAC ..(((..(((((.((((((((...))))))))))))))))..(((((((((.....((((.((.((((.((((....)))).)))).)).))))...)).....)))-))))........ ( -40.20) >consensus UUGGGCAUUGUCUAUUGCUCCUUUGGAGCGAUGACGACCCGUUUUGUGAGCCUUGUUCGUUUGUUUCAGCGGCCCGGGCCGAUGAAAUGCAUGAUCCGCCAUAGUUACCAGGUAAUUAAC ..(((..(((((.((((((((...))))))))))))))))...(((((.((.....((((.(((((((.((((....)))).))))))).))))...)))))))................ (-36.67 = -37.55 + 0.88)

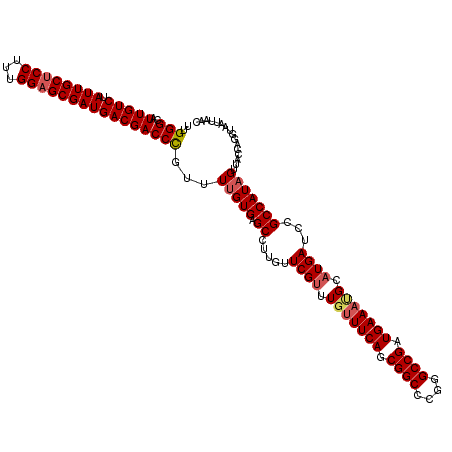
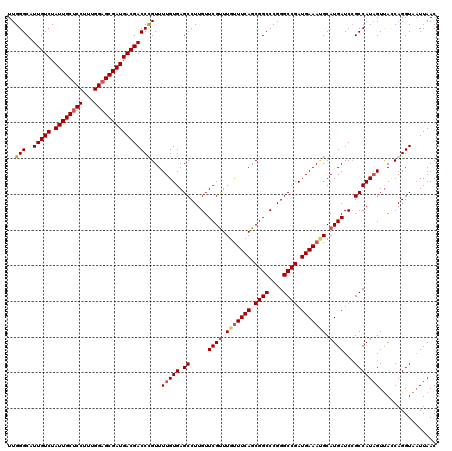
| Location | 21,269,225 – 21,269,345 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.64 |
| Mean single sequence MFE | -38.46 |
| Consensus MFE | -35.55 |
| Energy contribution | -35.67 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21269225 120 - 23771897 GUCCAAUCUUCCCUGUGAAAGGUGAACGCCCAUGUGGGUUAGAACUCUGGAUGAUGCUAUUAAGAUUUCCACAUACCUUUUUGGGCAUUGUCUAUUGCUCCUUUGGAGCGAUGACGACCC (((.....((((((.....))).))).(((((...((((..(((.(((.((((....)))).))).))).....))))...)))))...(((.((((((((...)))))))))))))).. ( -38.20) >DroSec_CAF1 27502 120 - 1 GUCCAAUCUUCCCUGUGAAAGGUGAAGGCACAUGUGGAUUAGAACUCUGGAGGAUGACAUUAAGAUUUCCACAUACCUUUUUGGGCAUUGUCUAUUGCUCCUUUGGAGCGAUGACGACCU ((((((((((((((.....))).)))))...(((((((((((....)))).(.....).........)))))))......)))))).(((((.((((((((...)))))))))))))... ( -39.60) >DroSim_CAF1 29758 120 - 1 GUCCAAUCUUCCCUGUGAAAGGUGAAGGCCCAUGUGGGUUAGAACUCUGGAGGAUGACAUUAAGAUUUCCACAUACCUUUUUGGGCAUUGUCUAUUGCCCCUUUGGAGCGAUGACGACCU (((....(((((((.....))).))))(((((...((((..(((.(((.((........)).))).))).....))))...)))))...(((.(((((((....)).))))))))))).. ( -35.70) >DroEre_CAF1 28677 120 - 1 GUCCAAUCUUUCCUGUGGUAGGUGAAGUGCCAUGUGGGUUAGAGCUCUGGAGGAUCCCUUCAAGAUUUCCACAUACCUUUUUGGGCAUUGUCUAUUGCUCCUUUGGAGCGAUGACGACCC ((((((..........(((((((.....))).((((((......((.((((((...))))))))...))))))))))...)))))).(((((.((((((((...)))))))))))))... ( -40.32) >consensus GUCCAAUCUUCCCUGUGAAAGGUGAAGGCCCAUGUGGGUUAGAACUCUGGAGGAUGACAUUAAGAUUUCCACAUACCUUUUUGGGCAUUGUCUAUUGCUCCUUUGGAGCGAUGACGACCC ((((((.(((((((.....))).))))....(((((((.......(((.((((....)))).)))..)))))))......)))))).(((((.((((((((...)))))))))))))... (-35.55 = -35.67 + 0.13)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:05:36 2006