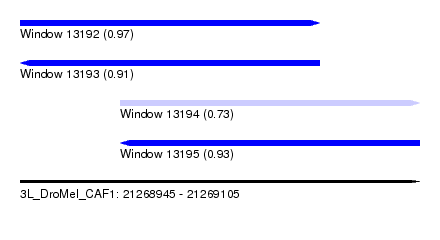
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,268,945 – 21,269,105 |
| Length | 160 |
| Max. P | 0.972766 |

| Location | 21,268,945 – 21,269,065 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.92 |
| Mean single sequence MFE | -32.35 |
| Consensus MFE | -24.89 |
| Energy contribution | -25.82 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21268945 120 + 23771897 AAUACAACAGUUGUUAGCAGCUGCUAACUCACAUACUCACCACUGCUGUUAAGCGCUACAGGCGAUGUUUUGCAAAACAUCGCCCAACAUCGAUAUCAAGAGAAACGUCGAGCUAUCGAG .........((.((((((....))))))..))...(((......(((....)))((....((((((((((....)))))))))).....(((((.((....))...)))))))....))) ( -32.60) >DroSec_CAF1 27233 120 + 1 AACACAACAGCUGUUAUCAGCUGCUUACUCAUAUACUCACCUCUGCUGUUAAGCGCUACAGACGAUGUUUUGCAAAAUAUCGCCCAACAUCGAUAUCAAGAGAAACGUCGUGUUAUCGAG (((((..((((((....)))))).................(((((((....)))........(((((((..((........))..)))))))......)))).......)))))...... ( -30.40) >DroSim_CAF1 29485 120 + 1 AACACAACAGCUGUUAUCAGCUGCUUACUCAUAUACUCACCUCUGCUGUUAAGCGCUACAGACGAUGUUUUGCAAAACAUCGCCCAACAUCGAUAUCAAGAGAAACGUCGUGUUAUCGAG (((((..((((((....)))))).................(((((((....)))........(((((((..((........))..)))))))......)))).......)))))...... ( -30.40) >DroEre_CAF1 28408 118 + 1 CA-AGUACCGCUGUUAGCAGCUGCGUUCCAACAUACUCACCACUGCUGUUAAGU-CUACAGGCGAUGUUUUGCAAAACAUCGCCUAACAUCGAUAUCAACAAUAACAUUGAGAUAUCGAG ..-.(((..(((..((((((.((.((........)).))...))))))...)))-.)))(((((((((((....)))))))))))....((((((((..((((...)))).)))))))). ( -36.00) >consensus AACACAACAGCUGUUAGCAGCUGCUUACUCACAUACUCACCACUGCUGUUAAGCGCUACAGACGAUGUUUUGCAAAACAUCGCCCAACAUCGAUAUCAAGAGAAACGUCGAGUUAUCGAG .......((((((....)))))).....................(((....)))......((((((((((....)))))))))).....((((((.((.((......)).)).)))))). (-24.89 = -25.82 + 0.94)



| Location | 21,268,945 – 21,269,065 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.92 |
| Mean single sequence MFE | -38.88 |
| Consensus MFE | -28.12 |
| Energy contribution | -29.25 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21268945 120 - 23771897 CUCGAUAGCUCGACGUUUCUCUUGAUAUCGAUGUUGGGCGAUGUUUUGCAAAACAUCGCCUGUAGCGCUUAACAGCAGUGGUGAGUAUGUGAGUUAGCAGCUGCUAACAACUGUUGUAUU ......(((((((..((......))..)))).((((((((((((((....)))))))))))..))))))..((((((((.((.(((((((......)))..)))).)).))))))))... ( -38.20) >DroSec_CAF1 27233 120 - 1 CUCGAUAACACGACGUUUCUCUUGAUAUCGAUGUUGGGCGAUAUUUUGCAAAACAUCGUCUGUAGCGCUUAACAGCAGAGGUGAGUAUAUGAGUAAGCAGCUGAUAACAGCUGUUGUGUU ......(((((((((((((((...(((((((((((..((((....))))..)))))))(((((.(.......).))))).....))))..))).))))(((((....))))))))))))) ( -38.90) >DroSim_CAF1 29485 120 - 1 CUCGAUAACACGACGUUUCUCUUGAUAUCGAUGUUGGGCGAUGUUUUGCAAAACAUCGUCUGUAGCGCUUAACAGCAGAGGUGAGUAUAUGAGUAAGCAGCUGAUAACAGCUGUUGUGUU ......(((((((((((((((...((((..((.(((((((((((((....))))))))))).....(((....))).)).))..))))..))).))))(((((....))))))))))))) ( -39.00) >DroEre_CAF1 28408 118 - 1 CUCGAUAUCUCAAUGUUAUUGUUGAUAUCGAUGUUAGGCGAUGUUUUGCAAAACAUCGCCUGUAG-ACUUAACAGCAGUGGUGAGUAUGUUGGAACGCAGCUGCUAACAGCGGUACU-UG .(((((((((((....((((((((.((.......((((((((((((....))))))))))))...-...)).)))))))).)))).)))))))......(((((.....)))))...-.. ( -39.42) >consensus CUCGAUAACACGACGUUUCUCUUGAUAUCGAUGUUGGGCGAUGUUUUGCAAAACAUCGCCUGUAGCGCUUAACAGCAGAGGUGAGUAUAUGAGUAAGCAGCUGAUAACAGCUGUUGUGUU .((((((.((.((......)).)).))))))....(((((((((((....))))))))))).....(((((.(......).))))).........((((((((....))))))))..... (-28.12 = -29.25 + 1.12)


| Location | 21,268,985 – 21,269,105 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.32 |
| Mean single sequence MFE | -29.51 |
| Consensus MFE | -19.83 |
| Energy contribution | -20.95 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21268985 120 + 23771897 CACUGCUGUUAAGCGCUACAGGCGAUGUUUUGCAAAACAUCGCCCAACAUCGAUAUCAAGAGAAACGUCGAGCUAUCGAGAAAAAUAUUGAAUCGACAGAACAUGUGCUCUAGCUCCAGC ..(((..(((((((((....((((((((((....)))))))))).....(((((.((((........((((....))))........)))))))))........))))).))))..))). ( -37.19) >DroSec_CAF1 27273 114 + 1 CUCUGCUGUUAAGCGCUACAGACGAUGUUUUGCAAAAUAUCGCCCAACAUCGAUAUCAAGAGAAACGUCGUGUUAUCGAGAAAAAUAUUAAAUCGACAGAACAUGUGCUCUAUC------ ....(((....)))((.(((.((((((((((.....((((((........)))))).....))))))))))(((.((((.............))))...))).)))))......------ ( -25.92) >DroSim_CAF1 29525 114 + 1 CUCUGCUGUUAAGCGCUACAGACGAUGUUUUGCAAAACAUCGCCCAACAUCGAUAUCAAGAGAAACGUCGUGUUAUCGAGAAAAAUAUUAAAUCGACAGAACAUGUGCUCUAUC------ (((((((....)))........(((((((..((........))..)))))))......))))....((((((((.((((.............))))...)))))).))......------ ( -24.72) >DroEre_CAF1 28447 113 + 1 CACUGCUGUUAAGU-CUACAGGCGAUGUUUUGCAAAACAUCGCCUAACAUCGAUAUCAACAAUAACAUUGAGAUAUCGAGAAAAACAUUGCAUCGUCAGAACAUGUGCUCUAGG------ ((((((((((...(-(...(((((((((((....)))))))))))....((((((((..((((...)))).))))))))))..))))..)))((....))....))).......------ ( -30.20) >consensus CACUGCUGUUAAGCGCUACAGACGAUGUUUUGCAAAACAUCGCCCAACAUCGAUAUCAAGAGAAACGUCGAGUUAUCGAGAAAAAUAUUAAAUCGACAGAACAUGUGCUCUAGC______ ...........(((((....((((((((((....)))))))))).....((((((.((.((......)).)).)))))).........................)))))........... (-19.83 = -20.95 + 1.12)



| Location | 21,268,985 – 21,269,105 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.32 |
| Mean single sequence MFE | -35.05 |
| Consensus MFE | -25.89 |
| Energy contribution | -27.20 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21268985 120 - 23771897 GCUGGAGCUAGAGCACAUGUUCUGUCGAUUCAAUAUUUUUCUCGAUAGCUCGACGUUUCUCUUGAUAUCGAUGUUGGGCGAUGUUUUGCAAAACAUCGCCUGUAGCGCUUAACAGCAGUG ((((((((.(((((....)))))((((((((((........((((....))))........)))).))))))((((((((((((((....)))))))))))..)))))))..)))).... ( -40.09) >DroSec_CAF1 27273 114 - 1 ------GAUAGAGCACAUGUUCUGUCGAUUUAAUAUUUUUCUCGAUAACACGACGUUUCUCUUGAUAUCGAUGUUGGGCGAUAUUUUGCAAAACAUCGUCUGUAGCGCUUAACAGCAGAG ------((((((((....))))))))..............(((((((.((.((......)).)).))))((((((..((((....))))..))))))(.((((........))))).))) ( -30.90) >DroSim_CAF1 29525 114 - 1 ------GAUAGAGCACAUGUUCUGUCGAUUUAAUAUUUUUCUCGAUAACACGACGUUUCUCUUGAUAUCGAUGUUGGGCGAUGUUUUGCAAAACAUCGUCUGUAGCGCUUAACAGCAGAG ------((((((((....))))))))...............((((((.((.((......)).)).)))))).((((((((((((((....)))))))))))..)))(((....))).... ( -33.00) >DroEre_CAF1 28447 113 - 1 ------CCUAGAGCACAUGUUCUGACGAUGCAAUGUUUUUCUCGAUAUCUCAAUGUUAUUGUUGAUAUCGAUGUUAGGCGAUGUUUUGCAAAACAUCGCCUGUAG-ACUUAACAGCAGUG ------..((((((....))))))....(((..((((..((((((((((.((((...))))..))))))))...((((((((((((....))))))))))))..)-)...)))))))... ( -36.20) >consensus ______GAUAGAGCACAUGUUCUGUCGAUUCAAUAUUUUUCUCGAUAACACGACGUUUCUCUUGAUAUCGAUGUUGGGCGAUGUUUUGCAAAACAUCGCCUGUAGCGCUUAACAGCAGAG ......((((((((....))))))))...............((((((.((.((......)).)).))))))(((((((((((((((....)))))))))))((........))))))... (-25.89 = -27.20 + 1.31)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:05:33 2006