| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,265,106 – 21,265,266 |
| Length | 160 |
| Max. P | 0.901332 |

| Location | 21,265,106 – 21,265,226 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.42 |
| Mean single sequence MFE | -32.27 |
| Consensus MFE | -29.70 |
| Energy contribution | -29.58 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21265106 120 + 23771897 GCGUUUGGCGUUUCCUUGACUUUAAAUUGUUGCCAUGUUUUCUAUAUUAAAUGUGUCGACUCUGCCAUUGUCUAACCGUGGCCUCGGGGAUGCACCUCAUCCCCAUAGAGUCACUCACAC .....(((((....................)))))................((((..(((((((((((.(.....).)))))...(((((((.....)))))))..))))))...)))). ( -35.05) >DroSec_CAF1 23224 120 + 1 GCGUUUGGCGUUUCCUUGACUUUAAAUUGUUGCCAUGUUUUCUAUAUUAAAUGUGUCGACUUUGCCAUUGUCUAACCGUGGCCUCGGGGAUGCACUCCCUCCCCAUAGAGUCACUCACAC ((.....)).......((((((((....((((.(((((((........))))))).))))...(((((.(.....).)))))...(((((.(.....).))))).))))))))....... ( -30.70) >DroSim_CAF1 24714 120 + 1 GCGUUUGGCGUUUCCUUGACUUUAAAUUGUUGCCAUGUUUUCUAUAUUAAAUGUGUCGACUCUGCCAUUGUCUAACCGUGGCCUCGGGGAUGCACUCCCUCCCCAUAGAGUCACUCACAC .....(((((....................)))))................((((..(((((((((((.(.....).)))))...(((((.(.....).)))))..))))))...)))). ( -30.95) >DroEre_CAF1 24316 120 + 1 GCGUUUGGCGUUUCCUUGACUUUAAAUUGUUGCCAUGUUUCCUAUAUUAAAUAUGUCGACUCUGCCAUUGUCUAACUGUGGCCUCGGGGAAGCACCACCUCCUCAUUGAGUCACUCACAC .....(((.((((((((((.........((((.(((((((........))))))).))))...(((((.(.....).))))).)))))))))).))).........((((...))))... ( -32.40) >consensus GCGUUUGGCGUUUCCUUGACUUUAAAUUGUUGCCAUGUUUUCUAUAUUAAAUGUGUCGACUCUGCCAUUGUCUAACCGUGGCCUCGGGGAUGCACCCCCUCCCCAUAGAGUCACUCACAC ((.....)).......((((((((....((((.(((((((........))))))).))))...(((((.(.....).)))))...(((((.(.....).))))).))))))))....... (-29.70 = -29.58 + -0.13)

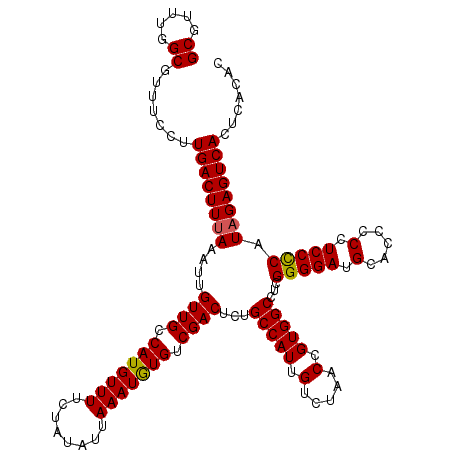
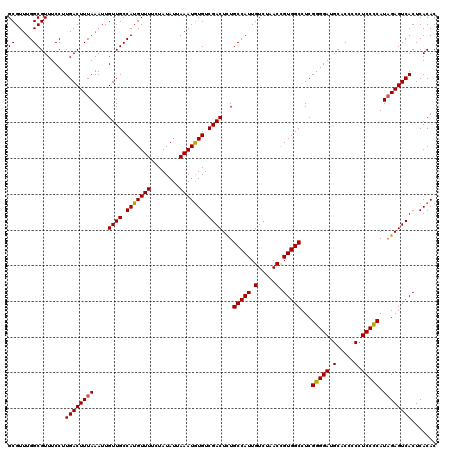
| Location | 21,265,106 – 21,265,226 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.42 |
| Mean single sequence MFE | -33.95 |
| Consensus MFE | -32.54 |
| Energy contribution | -33.47 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21265106 120 - 23771897 GUGUGAGUGACUCUAUGGGGAUGAGGUGCAUCCCCGAGGCCACGGUUAGACAAUGGCAGAGUCGACACAUUUAAUAUAGAAAACAUGGCAACAAUUUAAAGUCAAGGAAACGCCAAACGC (((((..(((((((.((((((((.....))))))))..((((..(.....)..))))))))))).)))))...............((((.((........))...(....)))))..... ( -39.20) >DroSec_CAF1 23224 120 - 1 GUGUGAGUGACUCUAUGGGGAGGGAGUGCAUCCCCGAGGCCACGGUUAGACAAUGGCAAAGUCGACACAUUUAAUAUAGAAAACAUGGCAACAAUUUAAAGUCAAGGAAACGCCAAACGC (((((..(((((((.((((((.........))))))))((((..(.....)..))))..))))).)))))...............((((.((........))...(....)))))..... ( -30.30) >DroSim_CAF1 24714 120 - 1 GUGUGAGUGACUCUAUGGGGAGGGAGUGCAUCCCCGAGGCCACGGUUAGACAAUGGCAGAGUCGACACAUUUAAUAUAGAAAACAUGGCAACAAUUUAAAGUCAAGGAAACGCCAAACGC (((((..(((((((.((((((.........))))))..((((..(.....)..))))))))))).)))))...............((((.((........))...(....)))))..... ( -35.00) >DroEre_CAF1 24316 120 - 1 GUGUGAGUGACUCAAUGAGGAGGUGGUGCUUCCCCGAGGCCACAGUUAGACAAUGGCAGAGUCGACAUAUUUAAUAUAGGAAACAUGGCAACAAUUUAAAGUCAAGGAAACGCCAAACGC ((((..(((((((..((.(((((.....))))).))..((((..(.....)..)))).)))))(((............(....).((....)).......)))..(....)))...)))) ( -31.30) >consensus GUGUGAGUGACUCUAUGGGGAGGGAGUGCAUCCCCGAGGCCACGGUUAGACAAUGGCAGAGUCGACACAUUUAAUAUAGAAAACAUGGCAACAAUUUAAAGUCAAGGAAACGCCAAACGC (((((..(((((((.((((((((.....))))))))..((((..(.....)..))))))))))).)))))...............((((.((........))...(....)))))..... (-32.54 = -33.47 + 0.94)

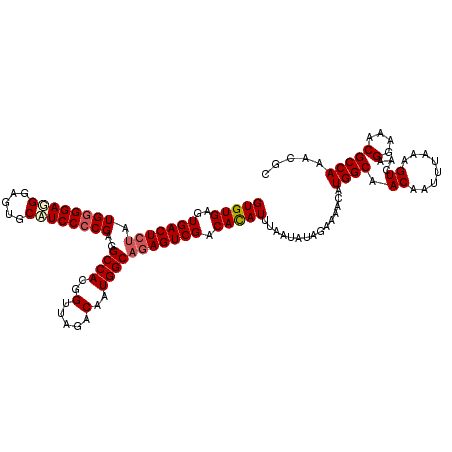
| Location | 21,265,146 – 21,265,266 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.92 |
| Mean single sequence MFE | -38.67 |
| Consensus MFE | -35.97 |
| Energy contribution | -35.73 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21265146 120 + 23771897 UCUAUAUUAAAUGUGUCGACUCUGCCAUUGUCUAACCGUGGCCUCGGGGAUGCACCUCAUCCCCAUAGAGUCACUCACACCCACUUCGCCGGCUUCCAAGUGGGGCAUUGGGGAAUGAAC ...........((((..(((((((((((.(.....).)))))...(((((((.....)))))))..))))))...))))((((...(.((.(((....))).)))...))))........ ( -41.60) >DroSec_CAF1 23264 120 + 1 UCUAUAUUAAAUGUGUCGACUUUGCCAUUGUCUAACCGUGGCCUCGGGGAUGCACUCCCUCCCCAUAGAGUCACUCACACCCACCUCGCCGGCUUCCAAGUGGGGCAUUGGGGAAUGAAC ...........((((..(((((((((((.(.....).)))))...(((((.(.....).)))))..))))))...))))((((((((((.((...))..))))))...))))........ ( -36.90) >DroSim_CAF1 24754 120 + 1 UCUAUAUUAAAUGUGUCGACUCUGCCAUUGUCUAACCGUGGCCUCGGGGAUGCACUCCCUCCCCAUAGAGUCACUCACACCCACCUCGCCGGCUUCCAAGUGGGGCAUUGGGGAAUGAAC ...........((((..(((((((((((.(.....).)))))...(((((.(.....).)))))..))))))...))))((((((((((.((...))..))))))...))))........ ( -39.50) >DroEre_CAF1 24356 120 + 1 CCUAUAUUAAAUAUGUCGACUCUGCCAUUGUCUAACUGUGGCCUCGGGGAAGCACCACCUCCUCAUUGAGUCACUCACACCCACCUCACCGGUUUCCAAGUGGGGCACUGGGGAUUGAGC .................(((((.(((((.(.....).)))))...(((((.(.....).)))))...))))).((((..((((((((((.((...))..))))))...))))...)))). ( -36.70) >consensus UCUAUAUUAAAUGUGUCGACUCUGCCAUUGUCUAACCGUGGCCUCGGGGAUGCACCCCCUCCCCAUAGAGUCACUCACACCCACCUCGCCGGCUUCCAAGUGGGGCAUUGGGGAAUGAAC ...........((((..(((((((((((.(.....).)))))...(((((.(.....).)))))..))))))...))))((((((((((.((...))..))))))...))))........ (-35.97 = -35.73 + -0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:05:27 2006