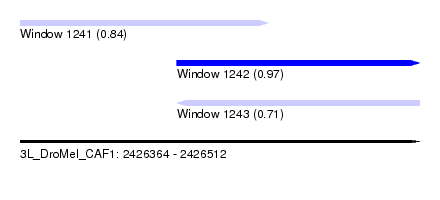
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,426,364 – 2,426,512 |
| Length | 148 |
| Max. P | 0.966951 |

| Location | 2,426,364 – 2,426,456 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 92.71 |
| Mean single sequence MFE | -33.13 |
| Consensus MFE | -30.93 |
| Energy contribution | -30.82 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
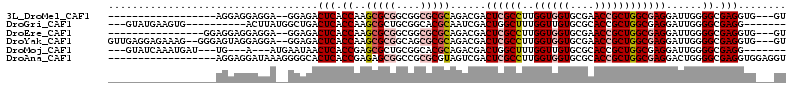
>3L_DroMel_CAF1 2426364 92 + 23771897 UGCGCCUCCGCAGGCCAUUGCCAGUCCGCUCCUGAGCGGCGGCAGCUGCUCCU---------CCUCGAC---CACCUCGCCCCAAUCCUCGCCAGCGGUUCGCA .(((...((((.((((((((((.(.(((((....))))))))))).)).....---------...(((.---....)))...........))).))))..))). ( -31.80) >DroSec_CAF1 59401 92 + 1 UGCGCCUCCGCAGGCCAUUGCCAGUCCGCUCAUGAGCGGCGGCAGCUGCUCCU---------CCUCGAC---CACCUCGCCCCAAUCCUCGCCAGCGGUUCGCA .(((...((((.((((((((((.(.(((((....))))))))))).)).....---------...(((.---....)))...........))).))))..))). ( -31.80) >DroSim_CAF1 53400 92 + 1 UGCGCCUCCGCAGGCCAUUGCCAGUCCGCUCCUGAGCGGCGGCAGCUGCUCCU---------CCUCGAC---CACCUCGCCCCAAUCCUCGCCAGCGGUUCGCA .(((...((((.((((((((((.(.(((((....))))))))))).)).....---------...(((.---....)))...........))).))))..))). ( -31.80) >DroEre_CAF1 53264 92 + 1 UGCGCCUCCGCAGGCCAUUGCCAGUCCGCUCCUGAGCGGCGGCAGCUGCUCCU---------CCUCGAC---CACCUCGCCCCAAUCCUCGCCAGCGGUUCGCA .(((...((((.((((((((((.(.(((((....))))))))))).)).....---------...(((.---....)))...........))).))))..))). ( -31.80) >DroYak_CAF1 54613 92 + 1 UGCGCCUCCGCAGGCCAUUGCCAGUCCGCUGCUGAGCGGCGGCAGCUGCUCCU---------CCUCGAC---CACCUCGCCCCAAUCCUCGCCAGCGGUUCGCA .(((...((((.(((....((((((.....)))).))((.(((((.((.((..---------....)).---)).)).))))).......))).))))..))). ( -32.50) >DroAna_CAF1 46581 104 + 1 UGCGCCUCCGCAGGCCACUGCCAGUCCUCUCCUGAGCGGCGGUAGCUGCACCUCGAACUCCGCCGCGACCUCCACCUCGCCCCAGUCCUCGCCAGCGGUGCGCA .((((..((((.(((.((((.(((.......))).(((((((.((...(.....)..)))))))))................))))....))).)))).)))). ( -39.10) >consensus UGCGCCUCCGCAGGCCAUUGCCAGUCCGCUCCUGAGCGGCGGCAGCUGCUCCU_________CCUCGAC___CACCUCGCCCCAAUCCUCGCCAGCGGUUCGCA .(((...((((.((((((((((.(.(((((....))))))))))).)).................(((........)))...........))).))))..))). (-30.93 = -30.82 + -0.11)

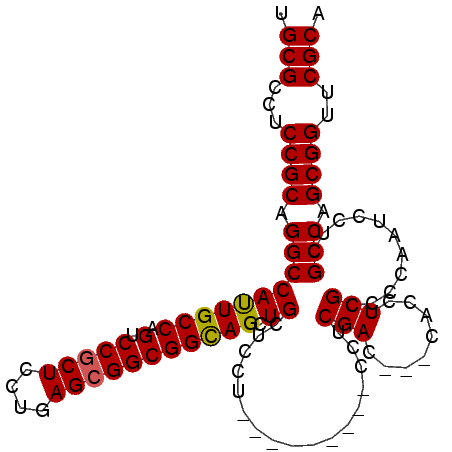
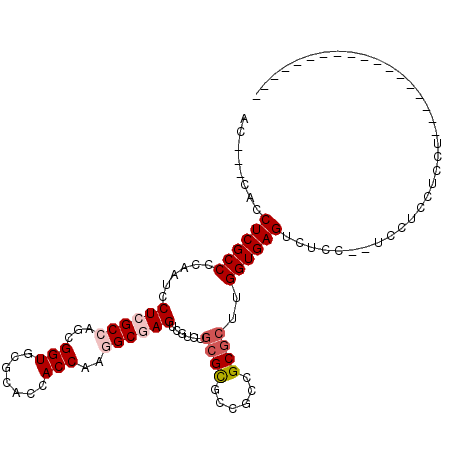
| Location | 2,426,422 – 2,426,512 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 72.45 |
| Mean single sequence MFE | -28.64 |
| Consensus MFE | -22.69 |
| Energy contribution | -23.47 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2426422 90 + 23771897 AC---CACCUCGCCCCAAUCCUCGCCAGCGGUUCGCACCACCAAGGCGAGUCGUCUGCGCGCCGCCGCGCUUGGUGAGUCUCC--UCCUCCUCCU------------------ ..---...((((((......((((((.(.(((....))).)...))))))......(((((....)))))..)))))).....--..........------------------ ( -28.90) >DroGri_CAF1 56262 93 + 1 -------CCUCGCCCCAAUCCUCGCCAGCGGUGCGCACAACCAAAGCCAGUCGAUUGCGUGCCGCAGCGCUUGGUGAGUCAGCCAUAAGU----------CACUUCAUAC--- -------.((((((........(((..((((..(((((.((........)).)..))))..)))).)))...))))))............----------..........--- ( -26.30) >DroEre_CAF1 53322 92 + 1 AC---CACCUCGCCCCAAUCCUCGCCAGCGGUUCGCACCACCAAGGCGAGUCGUCUGCGCGCCGCCGCGCUUGGUGAGUCUCC--UCCUCCUCCUCC---------------- ..---...((((((......((((((.(.(((....))).)...))))))......(((((....)))))..)))))).....--............---------------- ( -28.90) >DroYak_CAF1 54671 106 + 1 AC---CACCUCGCCCCAAUCCUCGCCAGCGGUUCGCACCACCAAGGCGAGUCGUCUGCGCGCUGCCGCGCUUGGUGAGUCUCC--UCCUCCUACUCCC--CUUUCUCCUCAAC ..---...((((((.........((..(((..((((.(......)))))..)))..))((((....))))..)))))).....--.............--............. ( -29.40) >DroMoj_CAF1 61009 94 + 1 -------CCUCGCCCCAAUCCUCGCCAGCGGUGCGCACAACCAAAGCCAGUCGUCUGCGUGCCGCAGCGCUCGGUGAGUUAUUCAU---U---CA---AUCAUUUGAUAC--- -------.............((((((((((.(((((((.......((.((....))))))).)))).)))).))))))........---(---((---(....))))...--- ( -27.91) >DroAna_CAF1 46648 95 + 1 ACCUCCACCUCGCCCCAGUCCUCGCCAGCGGUGCGCACCACCAAGGCGAGUCGACUACGCGCGGCCGCUCUCGGUGAGUGCCCCUUUAUCCUCCU------------------ .......((.(((...((((((((((...((((.....))))..))))))..))))..))).)).(((((.....)))))...............------------------ ( -30.40) >consensus AC___CACCUCGCCCCAAUCCUCGCCAGCGGUGCGCACCACCAAGGCGAGUCGUCUGCGCGCCGCCGCGCUUGGUGAGUCUCC__UCCUCCUCCU__________________ ........((((((......((((((...(((.......)))..))))))......((((......))))..))))))................................... (-22.69 = -23.47 + 0.78)
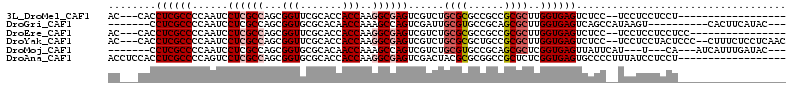
| Location | 2,426,422 – 2,426,512 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 72.45 |
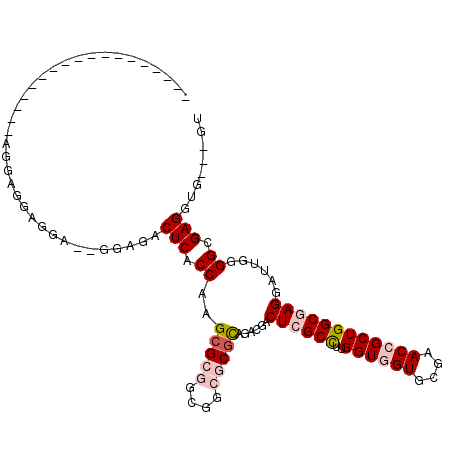
| Mean single sequence MFE | -35.77 |
| Consensus MFE | -27.23 |
| Energy contribution | -28.37 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.09 |
| Mean z-score | -0.71 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2426422 90 - 23771897 ------------------AGGAGGAGGA--GGAGACUCACCAAGCGCGGCGGCGCGCAGACGACUCGCCUUGGUGGUGCGAACCGCUGGCGAGGAUUGGGGCGAGGUG---GU ------------------........((--(....))).(((((((((....)))))......((((((..((((((....))))))))))))..)))).........---.. ( -38.80) >DroGri_CAF1 56262 93 - 1 ---GUAUGAAGUG----------ACUUAUGGCUGACUCACCAAGCGCUGCGGCACGCAAUCGACUGGCUUUGGUUGUGCGCACCGCUGGCGAGGAUUGGGGCGAGG------- ---..........----------............(((.((((.((((((((..((((..(((((......)))))))))..)))).))))....))))...))).------- ( -29.70) >DroEre_CAF1 53322 92 - 1 ----------------GGAGGAGGAGGA--GGAGACUCACCAAGCGCGGCGGCGCGCAGACGACUCGCCUUGGUGGUGCGAACCGCUGGCGAGGAUUGGGGCGAGGUG---GU ----------------..........((--(....))).(((((((((....)))))......((((((..((((((....))))))))))))..)))).........---.. ( -38.80) >DroYak_CAF1 54671 106 - 1 GUUGAGGAGAAAG--GGGAGUAGGAGGA--GGAGACUCACCAAGCGCGGCAGCGCGCAGACGACUCGCCUUGGUGGUGCGAACCGCUGGCGAGGAUUGGGGCGAGGUG---GU (((..........--...........((--(....))).(((((((((....)))))......((((((..((((((....))))))))))))..)))))))......---.. ( -39.50) >DroMoj_CAF1 61009 94 - 1 ---GUAUCAAAUGAU---UG---A---AUGAAUAACUCACCGAGCGCUGCGGCACGCAGACGACUGGCUUUGGUUGUGCGCACCGCUGGCGAGGAUUGGGGCGAGG------- ---...((((....)---))---)---........(((.((((.((((((((..(((..((((((......)))))))))..)))).))))....))))...))).------- ( -30.20) >DroAna_CAF1 46648 95 - 1 ------------------AGGAGGAUAAAGGGGCACUCACCGAGAGCGGCCGCGCGUAGUCGACUCGCCUUGGUGGUGCGCACCGCUGGCGAGGACUGGGGCGAGGUGGAGGU ------------------..(((............)))(((.....(.(((.(((.(((((..((((((..((((((....)))))))))))))))).).))).))).).))) ( -37.60) >consensus __________________AGGAGGAGGA__GGAGACUCACCAAGCGCGGCGGCGCGCAGACGACUCGCCUUGGUGGUGCGAACCGCUGGCGAGGAUUGGGGCGAGGUG___GU ...................................(((.((..(((((....)))))......((((((..((((((....))))))))))))......)).)))........ (-27.23 = -28.37 + 1.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:42 2006