
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,258,932 – 21,259,072 |
| Length | 140 |
| Max. P | 0.534454 |

| Location | 21,258,932 – 21,259,032 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 96.85 |
| Mean single sequence MFE | -27.35 |
| Consensus MFE | -23.26 |
| Energy contribution | -23.58 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.85 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
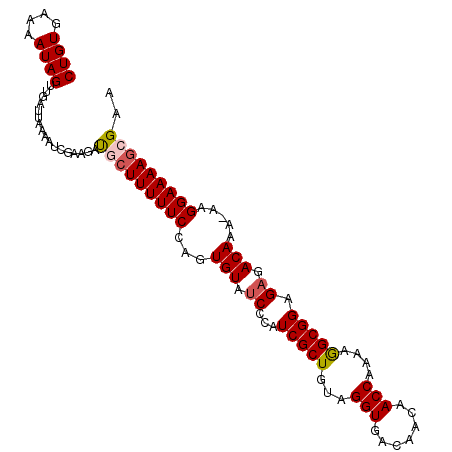
>3L_DroMel_CAF1 21258932 100 + 23771897 AACUUUGAACAAAUUUUGUGGAGUCCGGGAAAGGAAAUUUCGCUUUUCCUU-UUUGUCUCUCCGCCUUUUGGUUGUUGUCACCUACAGCGAUGGGGUACAC .(((((...((((....((((((....((((((((((.......)))))))-)))....))))))..))))((((((((.....))))))))))))).... ( -28.60) >DroSec_CAF1 16954 100 + 1 AACUUUGAACAAAUUUCGUGGAGUCCGGGAAAGGAAAUUUCGCUUUUCCUU-UUUGUCUCUCCGCCUUUUGGUUGUUGUCACCUACAGCGAUGGGAUACAC ............((..(((((((....((((((((((.......)))))))-)))....))))))......((((((((.....)))))))))..)).... ( -27.60) >DroSim_CAF1 16773 100 + 1 AACUUUGAACAAAUUUCGUGGAGUCCGGGAAAGGAAAUUUCGCUUUUCCUU-UUUGUCUCUCCGCCUUUUGGUUGUUGUCACCUACAGCGAUGGGAUACAC ............((..(((((((....((((((((((.......)))))))-)))....))))))......((((((((.....)))))))))..)).... ( -27.60) >DroEre_CAF1 18331 101 + 1 AACUUUGAACAAAUUUUGUGUACUCCGGGAAAGGAAAUUUCGCUUUUCCUUUUUUGUCUCUCCGCUUUUUGGUUGUUGUCACCUACAGCGAUGGGAUACAC .................(((((...((((((((((((.......))))))))))))((((..((((....(((.......)))...))))..))))))))) ( -25.60) >consensus AACUUUGAACAAAUUUCGUGGAGUCCGGGAAAGGAAAUUUCGCUUUUCCUU_UUUGUCUCUCCGCCUUUUGGUUGUUGUCACCUACAGCGAUGGGAUACAC .................((((((..((((.(((((((.......))))))).))))...))))))..(((.((((((((.....)))))))).)))..... (-23.26 = -23.58 + 0.31)



| Location | 21,258,970 – 21,259,072 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 84.88 |
| Mean single sequence MFE | -22.53 |
| Consensus MFE | -15.93 |
| Energy contribution | -16.80 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534454 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21258970 102 - 23771897 CUGUGAAAAUAGUUGAUUCAAAUCGAAAAUGCUUUUUCCAGUGUACCCCAUCGCUGUAGGUGACAACAACCAAAAGGCGGAGAGACAAA-AAGGAAAAGCGAA ((((....))))(((((....)))))...(((((((((...(((....(..((((...(((.......)))....))))..)..)))..-..))))))))).. ( -23.30) >DroSec_CAF1 16992 102 - 1 CUGUGGAAAUAGUUGAUUAAGAUCGAGGACUCUUUUUCCAGUGUAUCCCAUCGCUGUAGGUGACAACAACCAAAAGGCGGAGAGACAAA-AAGGAAAAGCGAA ((((....))))(((((....)))))....(((((((((..(((.((...(((((...(((.......)))....))))).)).)))..-..))))))).)). ( -21.20) >DroSim_CAF1 16811 102 - 1 CUGUGAAAAUAGUUGAUUAAGAUCGAGGAUGCUUUUUCCAGUGUAUCCCAUCGCUGUAGGUGACAACAACCAAAAGGCGGAGAGACAAA-AAGGAAAAGCGAA ((((....))))(((((....)))))...(((((((((...(((.((...(((((...(((.......)))....))))).)).)))..-..))))))))).. ( -24.40) >DroEre_CAF1 18369 87 - 1 -------------UGAUUGGAA---GACAUUGUUUUUCCAGUGUAUCCCAUCGCUGUAGGUGACAACAACCAAAAAGCGGAGAGACAAAAAAGGAAAAGCGAA -------------.........---....((((((((((..(((.((...(((((...(((.......)))....))))).)).))).....)))))))))). ( -21.20) >consensus CUGUGAAAAUAGUUGAUUAAAAUCGAAGAUGCUUUUUCCAGUGUAUCCCAUCGCUGUAGGUGACAACAACCAAAAGGCGGAGAGACAAA_AAGGAAAAGCGAA ((((....)))).................(((((((((...(((.((...(((((...(((.......)))....))))).)).))).....))))))))).. (-15.93 = -16.80 + 0.87)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:05:16 2006