| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,258,599 – 21,258,731 |
| Length | 132 |
| Max. P | 0.922626 |

| Location | 21,258,599 – 21,258,700 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 87.74 |
| Mean single sequence MFE | -22.88 |
| Consensus MFE | -15.77 |
| Energy contribution | -16.90 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563103 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
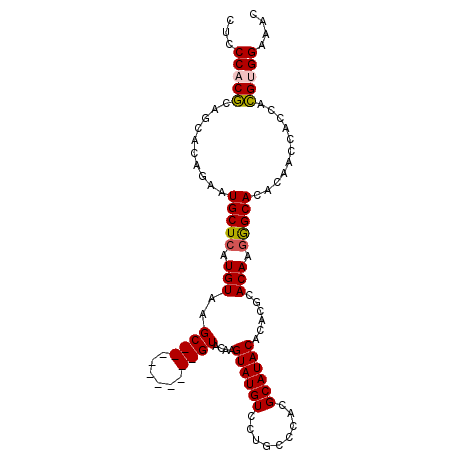
>3L_DroMel_CAF1 21258599 101 - 23771897 --------GUACAAGUAUGUCCUGUCCACGCAUACACACGCACAAGGGCACACAACCACCAAGAGGAAAGCAG-AACACACAGAUGUAAGGACACACUGACUUACAUUCA --------......((.(((.((((((..((........))....((........)).......))...))))-.))).)).((((((((..........)))))))).. ( -19.30) >DroSec_CAF1 16628 102 - 1 --------GUACAAGUAUGUCCUGCCCACGCAUACACACGCACAAGGGCACACAACCACCACGUGGAAACCAGGAGCACACAGAUGUAAGGACACACUGACUUACAUUCA --------......((.((((((((((..((........))....))))......((((...)))).....)))).)).)).((((((((..........)))))))).. ( -24.90) >DroSim_CAF1 16447 102 - 1 --------GUACAAGUAUGUCCUGCCCACGCAUACACACGCACAAGGGCACACAACCACCACGUGGAAACCAGGAGCACACAGAUGUAAGGACACACUGACUUACAUUCA --------......((.((((((((((..((........))....))))......((((...)))).....)))).)).)).((((((((..........)))))))).. ( -24.90) >DroEre_CAF1 17977 104 - 1 GUAAGUAUGUACGAGUAUGUCCUGCCCACGCAUACACACGCACAACAGCACACAACC---ACGGGGAAACCAGAA--ACACAGAUGUAAGG-CGCCCUGACUUACAUUCA ((((((.(((....((((((.........))))))....((......))..)))...---.(((((...((....--(((....)))..))-..)))))))))))..... ( -22.40) >consensus ________GUACAAGUAUGUCCUGCCCACGCAUACACACGCACAAGGGCACACAACCACCACGUGGAAACCAGGAGCACACAGAUGUAAGGACACACUGACUUACAUUCA ........(((..(((.((((((((((..((........))....))))......(((.....)))......................)))))).)))....)))..... (-15.77 = -16.90 + 1.13)


| Location | 21,258,638 – 21,258,731 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 87.41 |
| Mean single sequence MFE | -35.55 |
| Consensus MFE | -27.36 |
| Energy contribution | -28.42 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922626 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21258638 93 + 23771897 CUUUCCUCUUGGUGGUUGUGUGCCCUUGUGCGUGUGUAUGCGUGGACAGGACAUACUUGUAC----------GCUUACACGAGCAUUCUGUACUGAGUGGGAG ..((((.((..(((((.....)))...((((.((((((.((((..(((((.....)))))))----------)).)))))).)))).....))..)).)))). ( -32.70) >DroSec_CAF1 16668 93 + 1 GUUUCCACGUGGUGGUUGUGUGCCCUUGUGCGUGUGUAUGCGUGGGCAGGACAUACUUGUAC----------GCUUACAUGAGCAUUCUGUGCUGCGUGGGAG ..(((((((..(((((.....)))...((((..(((((.((((..(((((.....)))))))----------)).)))))..)))).....))..))))))). ( -36.10) >DroSim_CAF1 16487 93 + 1 GUUUCCACGUGGUGGUUGUGUGCCCUUGUGCGUGUGUAUGCGUGGGCAGGACAUACUUGUAC----------GCUUACAUGAGCAUUCUGUGCUGCGUGGGAG ..(((((((..(((((.....)))...((((..(((((.((((..(((((.....)))))))----------)).)))))..)))).....))..))))))). ( -36.10) >DroEre_CAF1 18014 100 + 1 GUUUCCCCGU---GGUUGUGUGCUGUUGUGCGUGUGUAUGCGUGGGCAGGACAUACUCGUACAUACUUACAUGCUUACAUGAGCAUUCUGAGCUGCGUGGGAG ...((((((.---.(((..(((((..((((((((((((((..((((.........))))..)))))..))))))..)))..)))))....)))..)).)))). ( -37.30) >consensus GUUUCCACGUGGUGGUUGUGUGCCCUUGUGCGUGUGUAUGCGUGGGCAGGACAUACUUGUAC__________GCUUACAUGAGCAUUCUGUGCUGCGUGGGAG ..(((((((((((....(((((((((..(((((....)))))..))..)).)))))................((((....)))).......))))))))))). (-27.36 = -28.42 + 1.06)

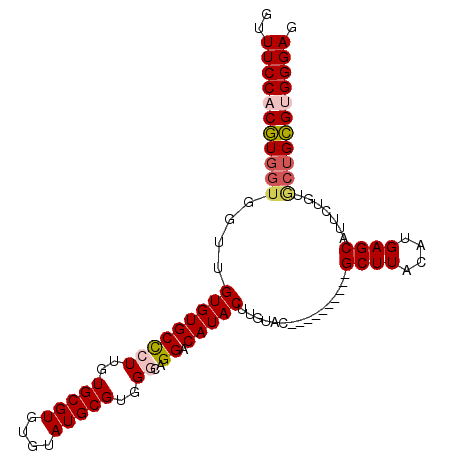
| Location | 21,258,638 – 21,258,731 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 87.41 |
| Mean single sequence MFE | -22.03 |
| Consensus MFE | -14.86 |
| Energy contribution | -15.43 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21258638 93 - 23771897 CUCCCACUCAGUACAGAAUGCUCGUGUAAGC----------GUACAAGUAUGUCCUGUCCACGCAUACACACGCACAAGGGCACACAACCACCAAGAGGAAAG ......(((.........(((..(((((.((----------(((((.(.....).)))..)))).)))))..)))...((........)).....)))..... ( -19.70) >DroSec_CAF1 16668 93 - 1 CUCCCACGCAGCACAGAAUGCUCAUGUAAGC----------GUACAAGUAUGUCCUGCCCACGCAUACACACGCACAAGGGCACACAACCACCACGUGGAAAC ...(((((..........(((((.(((..((----------((....((((((.........))))))..))))))).)))))...........))))).... ( -23.10) >DroSim_CAF1 16487 93 - 1 CUCCCACGCAGCACAGAAUGCUCAUGUAAGC----------GUACAAGUAUGUCCUGCCCACGCAUACACACGCACAAGGGCACACAACCACCACGUGGAAAC ...(((((..........(((((.(((..((----------((....((((((.........))))))..))))))).)))))...........))))).... ( -23.10) >DroEre_CAF1 18014 100 - 1 CUCCCACGCAGCUCAGAAUGCUCAUGUAAGCAUGUAAGUAUGUACGAGUAUGUCCUGCCCACGCAUACACACGCACAACAGCACACAACC---ACGGGGAAAC .((((.((..((((...(((((((((....))))..)))))....)))).(((.(((.....((........))....))).))).....---.))))))... ( -22.20) >consensus CUCCCACGCAGCACAGAAUGCUCAUGUAAGC__________GUACAAGUAUGUCCUGCCCACGCAUACACACGCACAAGGGCACACAACCACCACGUGGAAAC ...(((((..........(((((.(((..((((......))))....((((((.........))))))......))).)))))...........))))).... (-14.86 = -15.43 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:05:14 2006