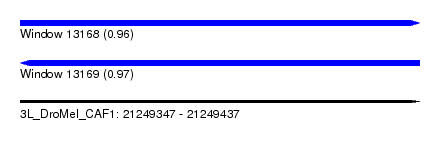
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,249,347 – 21,249,437 |
| Length | 90 |
| Max. P | 0.972353 |

| Location | 21,249,347 – 21,249,437 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 79.66 |
| Mean single sequence MFE | -29.83 |
| Consensus MFE | -18.02 |
| Energy contribution | -19.85 |
| Covariance contribution | 1.83 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961364 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
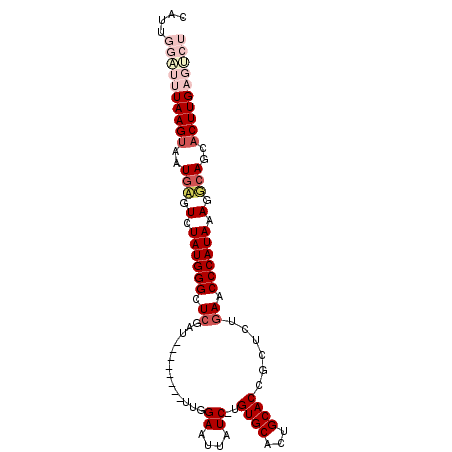
>3L_DroMel_CAF1 21249347 90 + 23771897 CAUAGGAUUUAAGUAAUGAGUCUAUGGGCUCGAU--------UUGGAAUUAUC-UGUGCACUGCACCGCUCUGAACCCAUAAAGGCAGCACUUGAGUCU ....((((((((((..((..(.((((((.(((((--------........)))-.((((...))))......)).)))))).)..))..)))))))))) ( -25.20) >DroPse_CAF1 6762 99 + 1 UCUUUGGGAUAAGUGCUGCAUCUAUGGGUUCCAUGCAUUAUUAUGGAAUUAUCUUGUGCACUGCACCGUACAGAACCCAUACAGGCAGCACUUGGGGAU .........((((((((((...(((((((((((((......))))))....(((.((((........))))))))))))))...))))))))))..... ( -37.10) >DroSec_CAF1 7422 90 + 1 CAUUGGAUUUAAGUAAUGAGUCUAUGGGCUCGAU--------UUGGAAUUAUC-UGUGCACUGCACAGCUCUAAACCCAUAAAGUCAGCUCUUGAGUCU ....(((((((((...(((.(.((((((.....(--------(((((.....(-(((((...)))))).)))))))))))).).)))...))))))))) ( -29.20) >DroSim_CAF1 7454 90 + 1 CAUUGGAUUUAAGUAAUGAGUCUAUGGGUUCGAU--------UUGGAAUUAUC-CGUGCACUGCACAGCUCUGAACCCAUAAAGUCAGCACUUGAGUCU ....((((((((((..(((.(.((((((((((..--------..(((....))-)((((...)))).....)))))))))).).)))..)))))))))) ( -35.30) >DroEre_CAF1 7370 90 + 1 CAUAGGGUUUAAGUAAUAAGUCUAUGGGCUCGAU--------UUGGAAUUAUC-UGUGCACUGCACCGCUCUGAACCCAUAAAGUCAGCACUUGAGCCU ....((((((((((........((((((.(((((--------........)))-.((((...))))......)).))))))........)))))))))) ( -23.89) >DroPer_CAF1 6773 99 + 1 UCUUUGGGAUAAGUAUUGCAUUUAUGGGUUCCAUGCAUUAUUAUGGAAUUGUCUUGUGCACUGCACCGUACAGAACCCAUACAGGCAGCACUUGGGGGU .........(((((.((((...(((((((((((((......))))))....(((.((((........))))))))))))))...)))).)))))..... ( -28.30) >consensus CAUUGGAUUUAAGUAAUGAGUCUAUGGGCUCGAU________UUGGAAUUAUC_UGUGCACUGCACCGCUCUGAACCCAUAAAGGCAGCACUUGAGUCU ....((((((((((..(((.(.((((((.((..............((....))..((((...))))......)).)))))).).)))..)))))))))) (-18.02 = -19.85 + 1.83)


| Location | 21,249,347 – 21,249,437 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 79.66 |
| Mean single sequence MFE | -28.47 |
| Consensus MFE | -24.53 |
| Energy contribution | -24.20 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
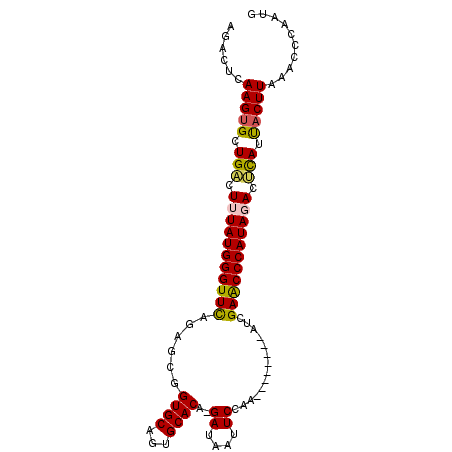
>3L_DroMel_CAF1 21249347 90 - 23771897 AGACUCAAGUGCUGCCUUUAUGGGUUCAGAGCGGUGCAGUGCACA-GAUAAUUCCAA--------AUCGAGCCCAUAGACUCAUUACUUAAAUCCUAUG ......(((((.((..(((((((((((......((((...)))).-(((........--------))))))))))))))..)).))))).......... ( -25.40) >DroPse_CAF1 6762 99 - 1 AUCCCCAAGUGCUGCCUGUAUGGGUUCUGUACGGUGCAGUGCACAAGAUAAUUCCAUAAUAAUGCAUGGAACCCAUAGAUGCAGCACUUAUCCCAAAGA ......(((((((((.(.((((((((((((.((((((...))))..((....))........)).)))))))))))).).))))))))).......... ( -36.80) >DroSec_CAF1 7422 90 - 1 AGACUCAAGAGCUGACUUUAUGGGUUUAGAGCUGUGCAGUGCACA-GAUAAUUCCAA--------AUCGAGCCCAUAGACUCAUUACUUAAAUCCAAUG ......(((...(((.(((((((((((.((.((((((...)))))-)..........--------.))))))))))))).)))...))).......... ( -23.00) >DroSim_CAF1 7454 90 - 1 AGACUCAAGUGCUGACUUUAUGGGUUCAGAGCUGUGCAGUGCACG-GAUAAUUCCAA--------AUCGAACCCAUAGACUCAUUACUUAAAUCCAAUG ......(((((.(((.(((((((((((......((((...))))(-((....)))..--------...))))))))))).))).))))).......... ( -30.60) >DroEre_CAF1 7370 90 - 1 AGGCUCAAGUGCUGACUUUAUGGGUUCAGAGCGGUGCAGUGCACA-GAUAAUUCCAA--------AUCGAGCCCAUAGACUUAUUACUUAAACCCUAUG (((...(((((.(((.(((((((((((......((((...)))).-(((........--------)))))))))))))).))).)))))....)))... ( -27.20) >DroPer_CAF1 6773 99 - 1 ACCCCCAAGUGCUGCCUGUAUGGGUUCUGUACGGUGCAGUGCACAAGACAAUUCCAUAAUAAUGCAUGGAACCCAUAAAUGCAAUACUUAUCCCAAAGA ......(((((.(((...((((((((((((...((((...))))...)))....(((........))))))))))))...))).))))).......... ( -27.80) >consensus AGACUCAAGUGCUGACUUUAUGGGUUCAGAGCGGUGCAGUGCACA_GAUAAUUCCAA________AUCGAACCCAUAGACUCAUUACUUAAACCCAAUG ......(((((.(((.(((((((((((......((((...))))..((....))..............))))))))))).))).))))).......... (-24.53 = -24.20 + -0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:05:08 2006