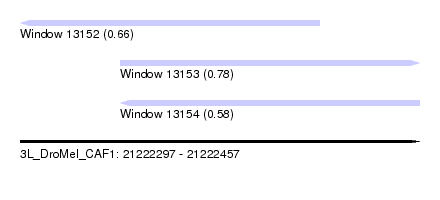
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,222,297 – 21,222,457 |
| Length | 160 |
| Max. P | 0.778542 |

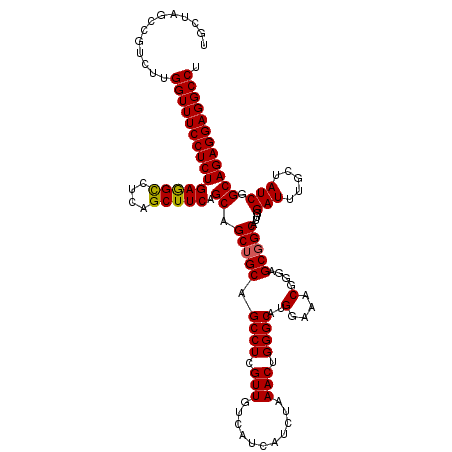
| Location | 21,222,297 – 21,222,417 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -46.60 |
| Consensus MFE | -41.54 |
| Energy contribution | -41.35 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21222297 120 - 23771897 UGCUAGCCGUCUUGGUUUCCUCUGAGGCCUCAGCUUCAGCAGCUGCAGCCUCGUUGUCAUCAUCUAAACUGGGCAUGGAAACCGGUGCGGCAUUGGAUUUGCUAUCGGCAGAGGAGGCCU .(((.(((((.(((((((((...(((((..(((((.....)))))..)))))..((((.............)))).))))))))).)))))......(((((.....)))))...))).. ( -46.72) >DroSim_CAF1 89338 120 - 1 UGCUAGCCGCCUUGGUUUCCUCUGAGGCCUCAGCUUCAGCAGCUGCAGCCUCGUUGUCAUCAUCUAAACUGGGCAUGGAAACAGGCGCGGCAUUGGAUUUGCUAUCGGCAGAGGAGGCCU .............(((((((((((((((..(((((.....)))))..)))))((((.((..((((((.(((.((.((....)).)).)))..)))))).))....)))))))))))))). ( -49.00) >DroEre_CAF1 41721 120 - 1 UGCUAGCCGUCUUGGUUUCCUCUGAGGUCUCAGCUUCAGCAGCGGCAGCCUCGUUGUCAUCAUCUAAACUGGGCAUGGAAACGGGAGCGGCGUUGGAUUUGCUAUCGGCAGAGGAGGCCU .............(((((((((((.(((..(((.((((((.((.((.((((.(((...........))).)))).((....))...)).)))))))).)))..)))..))))))))))). ( -45.80) >DroYak_CAF1 146876 120 - 1 UGCUAGCCGUCUUGGUUUCCUCUGAAGUCUCAGCUUCAGCAGCUGCAGCCUCGUUGUCAUCAUCUAAACUGGGCAUGGAAACGGGAGCGGCGUUGGAUUUGCUAUCAGCAGAGGAGGCCU .............(((((((((((..((..(((.((((((.(((((.((((.(((...........))).)))).((....))...))))))))))).)))..))...))))))))))). ( -44.90) >consensus UGCUAGCCGUCUUGGUUUCCUCUGAGGCCUCAGCUUCAGCAGCUGCAGCCUCGUUGUCAUCAUCUAAACUGGGCAUGGAAACGGGAGCGGCAUUGGAUUUGCUAUCGGCAGAGGAGGCCU .............(((((((((((((((....))))).((.(((((.((((.(((...........))).))))..(....)....)))))....(((.....))).)))))))))))). (-41.54 = -41.35 + -0.19)


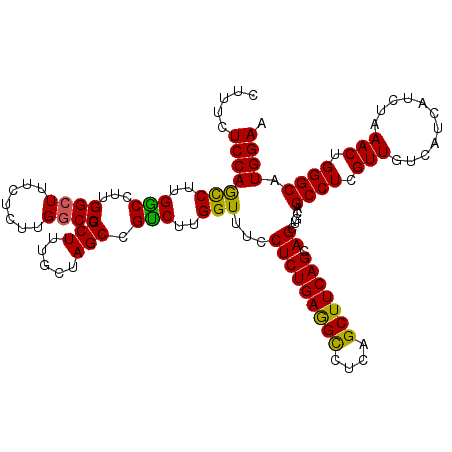
| Location | 21,222,337 – 21,222,457 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -37.48 |
| Consensus MFE | -34.31 |
| Energy contribution | -34.38 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21222337 120 + 23771897 UUCCAUGCCCAGUUUAGAUGAUGACAACGAGGCUGCAGCUGCUGAAGCUGAGGCCUCAGAGGAAACCAAGACGGCUAGCAAAGCAGGUCAAGAGAAAGCCAAGGCCAAGGCUGGAGAAAG (((((.(((..((((.............((((((.(((((.....))))).)))))).(.(....)).))))((((.((...)).(((.........)))..))))..)))))))).... ( -40.70) >DroSim_CAF1 89378 120 + 1 UUCCAUGCCCAGUUUAGAUGAUGACAACGAGGCUGCAGCUGCUGAAGCUGAGGCCUCAGAGGAAACCAAGGCGGCUAGCAAAGCAGGCCAAGAGAAAGCCAAGGCCAAGGUUGGAGAAAG ((((....((.(((........)))...((((((.(((((.....))))).))))))...)).((((..(((.(((.....))).(((.........)))...)))..)))))))).... ( -40.70) >DroEre_CAF1 41761 120 + 1 UUCCAUGCCCAGUUUAGAUGAUGACAACGAGGCUGCCGCUGCUGAAGCUGAGACCUCAGAGGAAACCAAGACGGCUAGCAAAGCGGGCCAAGAGAAAGCCAAGGCCAAGACUGGAGAAAG ........(((((((............((..(((((((((.......((((....)))).(....)..)).)))).)))....))((((.............)))).)))))))...... ( -33.52) >DroYak_CAF1 146916 120 + 1 UUCCAUGCCCAGUUUAGAUGAUGACAACGAGGCUGCAGCUGCUGAAGCUGAGACUUCAGAGGAAACCAAGACGGCUAGCAAAGCAGGCCAAGAGAAAGCCAAGGUCAAGGCUGGAGAAAG (((((.(((((.......)).((((.....((((.(((((.....)))))....(((.(.(....)).....((((.((...)).))))....)))))))...)))).)))))))).... ( -35.00) >consensus UUCCAUGCCCAGUUUAGAUGAUGACAACGAGGCUGCAGCUGCUGAAGCUGAGACCUCAGAGGAAACCAAGACGGCUAGCAAAGCAGGCCAAGAGAAAGCCAAGGCCAAGGCUGGAGAAAG (((((.(((..((((.............((((((.(((((.....))))).)))))).(.(....)).))))((((.((...)).(((.........)))..))))..)))))))).... (-34.31 = -34.38 + 0.06)



| Location | 21,222,337 – 21,222,457 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -35.22 |
| Consensus MFE | -31.91 |
| Energy contribution | -30.98 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575092 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21222337 120 - 23771897 CUUUCUCCAGCCUUGGCCUUGGCUUUCUCUUGACCUGCUUUGCUAGCCGUCUUGGUUUCCUCUGAGGCCUCAGCUUCAGCAGCUGCAGCCUCGUUGUCAUCAUCUAAACUGGGCAUGGAA .....((((((((.(((....)).......((((..((...(..((((.....))))..)...(((((..(((((.....)))))..))))))).)))).........).)))).)))). ( -35.70) >DroSim_CAF1 89378 120 - 1 CUUUCUCCAACCUUGGCCUUGGCUUUCUCUUGGCCUGCUUUGCUAGCCGCCUUGGUUUCCUCUGAGGCCUCAGCUUCAGCAGCUGCAGCCUCGUUGUCAUCAUCUAAACUGGGCAUGGAA .....((((.....((((..((.....))..))))(((.((((((((.((((..(......)..))))....)))..)))))..)))((((.(((...........))).)))).)))). ( -37.80) >DroEre_CAF1 41761 120 - 1 CUUUCUCCAGUCUUGGCCUUGGCUUUCUCUUGGCCCGCUUUGCUAGCCGUCUUGGUUUCCUCUGAGGUCUCAGCUUCAGCAGCGGCAGCCUCGUUGUCAUCAUCUAAACUGGGCAUGGAA .....(((((((..((((..((.....))..)))).(((..(..((((.....))))..).(((((((....))))))).)))))).((((.(((...........))).)))).)))). ( -33.60) >DroYak_CAF1 146916 120 - 1 CUUUCUCCAGCCUUGACCUUGGCUUUCUCUUGGCCUGCUUUGCUAGCCGUCUUGGUUUCCUCUGAAGUCUCAGCUUCAGCAGCUGCAGCCUCGUUGUCAUCAUCUAAACUGGGCAUGGAA .....((((((((.((...((((.....(..(((.(((...(((((((.....))))....(((((((....))))))).))).))))))..)..))))...))......)))).)))). ( -33.80) >consensus CUUUCUCCAGCCUUGGCCUUGGCUUUCUCUUGGCCUGCUUUGCUAGCCGUCUUGGUUUCCUCUGAGGCCUCAGCUUCAGCAGCUGCAGCCUCGUUGUCAUCAUCUAAACUGGGCAUGGAA .....(((((((..(((...((((.......)))).(((.....))).)))..)))...(((((((((....))))))).)).....((((.(((...........))).)))).)))). (-31.91 = -30.98 + -0.94)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:04:53 2006