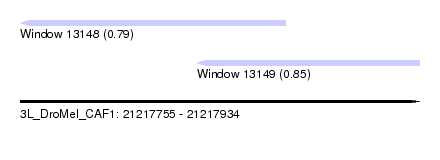
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,217,755 – 21,217,934 |
| Length | 179 |
| Max. P | 0.853270 |

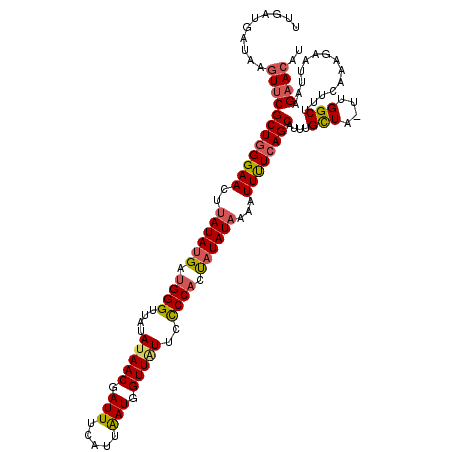
| Location | 21,217,755 – 21,217,874 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.01 |
| Mean single sequence MFE | -25.87 |
| Consensus MFE | -22.54 |
| Energy contribution | -23.67 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21217755 119 - 23771897 UUGAUGAUAAGUUCGCUGGGAACUUAUAUGAUGGGUUAUAUAACGAUUUCAUUAAUGGUUCUUUCCCACCAUAUACAAUUUUCAGCAUUUGCUA-UUGGCUUUCAAAGAAUUAAGCACAU .((.((...(((((((((..((..((((((.((((......(((.(((.....))).)))....)))).))))))...))..))))....((..-...)).......)))))...)))). ( -27.30) >DroSim_CAF1 84818 119 - 1 UUGAUGAUAAGUUCGCUGGGAACUUAUAUGAUGGGUUAUAUAACGAUUUCAUUAAUGGUUGUUCCCCACUAUAUAAAAUUUUCAGCAUUUGCUA-UUGGCAUUCAAAGAAUUAAGAACAU ..........((((((((..((.(((((((.((((....(((((.(((.....))).)))))..)))).)))))))..))..))))...(((..-...))).............)))).. ( -31.30) >DroEre_CAF1 37203 119 - 1 CUAAUGAUAAAUUCGCUUGGAACUUAUAUGACGGGUUAUAUAACGAUUUCAU-GAUGGUUAUACUCCACUUUAUUACAUUCUUAGCAGUUGUUGGCAGGCUUUCUAAAACUUAAGAAAAU ((..((((((....(((.((((.....((((.(((...((((((.(((....-))).))))))..)).).))))....)))).)))..))))))..))..(((((........))))).. ( -19.00) >consensus UUGAUGAUAAGUUCGCUGGGAACUUAUAUGAUGGGUUAUAUAACGAUUUCAUUAAUGGUUAUUCCCCACUAUAUAAAAUUUUCAGCAUUUGCUA_UUGGCUUUCAAAGAAUUAAGAACAU ..........((((((((((((..((((((.((((....(((((.(((.....))).)))))..)))).))))))...))))))))....(((....)))..............)))).. (-22.54 = -23.67 + 1.12)

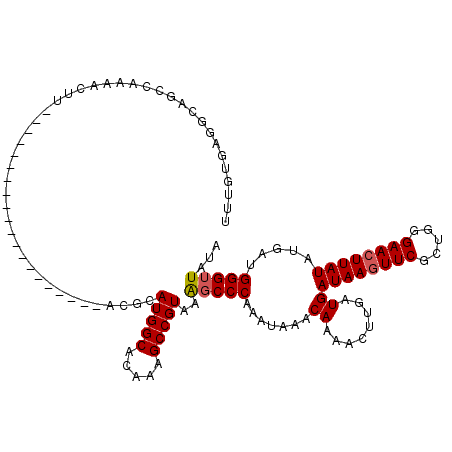
| Location | 21,217,834 – 21,217,934 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.00 |
| Mean single sequence MFE | -25.18 |
| Consensus MFE | -16.29 |
| Energy contribution | -16.73 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21217834 100 - 23771897 UUUGUGAGGCAUCCAAAACUU--------------------ACGCAUGGCACACAGCCGUAAAGCCCAAAUAAACAAAACUUGAUGAUAAGUUCGCUGGGAACUUAUAUGAUGGGUUAUA ...(((((..........)))--------------------))..(((((.....)))))..((((((......((.....))...((((((((.....))))))))....))))))... ( -24.30) >DroSim_CAF1 84897 100 - 1 UUUGUGAGGCAGCCAAAACUU--------------------ACGCAUGGCACAAAGCCGUAAAGCCCAAAUAAACAAAACUUGAUGAUAAGUUCGCUGGGAACUUAUAUGAUGGGUUAUA ...(((((..........)))--------------------))..(((((.....)))))..((((((......((.....))...((((((((.....))))))))....))))))... ( -24.30) >DroEre_CAF1 37282 120 - 1 UUUAUCAAGCAGCCAAAACUGUUGAUAAAAUCAAAAGGCCCUCGCAUGGCACAAAGCCGUAAGUCCCAAAUAAACAAAACCUAAUGAUAAAUUCGCUUGGAACUUAUAUGACGGGUUAUA ((((((((.(((......))))))))))).......((((((((...(((.....)))((((((.((((...........................)))).)))))).))).)))))... ( -26.93) >consensus UUUGUGAGGCAGCCAAAACUU____________________ACGCAUGGCACAAAGCCGUAAAGCCCAAAUAAACAAAACUUGAUGAUAAGUUCGCUGGGAACUUAUAUGAUGGGUUAUA .............................................(((((.....)))))..(((((.......((........))((((((((.....)))))))).....)))))... (-16.29 = -16.73 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:04:48 2006