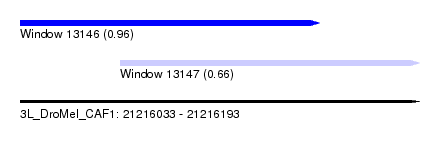
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,216,033 – 21,216,193 |
| Length | 160 |
| Max. P | 0.964856 |

| Location | 21,216,033 – 21,216,153 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.08 |
| Mean single sequence MFE | -51.43 |
| Consensus MFE | -44.30 |
| Energy contribution | -45.18 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
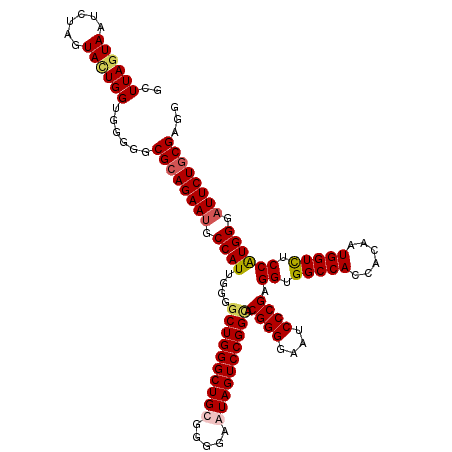
>3L_DroMel_CAF1 21216033 120 + 23771897 GCUUAGUAAUCUAGUACUGGUUGUGGCGCAGAACGCCAUUGGGACUGGGCUGCGGGGAGUAGUCCGGCACGGGGAAUCCCGAGGUGGCCACCACAAUGGUCUCCAUGGGAUUCUGCGAAG ..((((((......))))))..((((((.....))))))...(.(((((((((.....)))))))))).((.(((((((((.((.(((((......))))).)).))))))))).))... ( -52.90) >DroSim_CAF1 83113 120 + 1 GCUUAGUAAUCUAGUACUGGUGUUGGCGCAGAAUGCCAUUGGGGCUGGGCUGCGGGGAGUAGUCCGGCACGGGGAAUCCCGAGGUGGCCACCACUAUGGUCUCCAUGGGAUUCUGCGAGG (((.((((......))))))).....((((((((.((((....((((((((((.....)))))))))).((((....)))).((.(((((......))))).)))))).))))))))... ( -54.50) >DroEre_CAF1 35514 120 + 1 GCUUAAUAAUCUAGUACUGGUUGGUGCGCAGAAUGCCAUUCGGGCUGGGCUGCGGGGAAUAGUCCGGUACGGGGAAUCCCGAGGUGGCCACCACAAUGGUUUCCAUGGGAUUCUGCGAGG .(((.........((((((((((.(.(((((...(((.....)))....))))).)...))).)))))))(.(((((((((.((..((((......))))..)).))))))))).)))). ( -49.70) >DroYak_CAF1 142273 120 + 1 GCUUAGUAAUCUAGUAUUGGUGGGGACGAAGAAUGCCAUUGGGGCUGGGCUGCGGGGAAUAGUCCGGUACGGGGAAUCCCGAGGUGGCCACCACAAUGGUCUCCGUGGGAUUCUGCGACG .....(((..(((((.(..((((.(........).))))..).)))))..)))..(((....))).((.((.(((((((((.((.(((((......))))).)).))))))))).)))). ( -48.60) >consensus GCUUAGUAAUCUAGUACUGGUGGGGGCGCAGAAUGCCAUUGGGGCUGGGCUGCGGGGAAUAGUCCGGCACGGGGAAUCCCGAGGUGGCCACCACAAUGGUCUCCAUGGGAUUCUGCGAGG ..((((((......))))))......((((((((.((((....((((((((((.....)))))))))).((((....)))).((.(((((......))))).)))))).))))))))... (-44.30 = -45.18 + 0.88)

| Location | 21,216,073 – 21,216,193 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.19 |
| Mean single sequence MFE | -54.60 |
| Consensus MFE | -52.72 |
| Energy contribution | -52.10 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663918 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
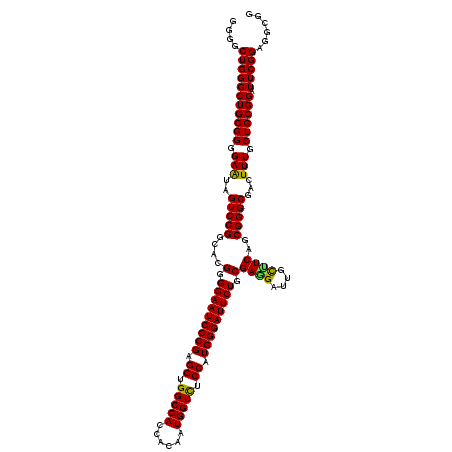
>3L_DroMel_CAF1 21216073 120 + 23771897 GGGACUGGGCUGCGGGGAGUAGUCCGGCACGGGGAAUCCCGAGGUGGCCACCACAAUGGUCUCCAUGGGAUUCUGCGAAGAUUGCUUCAGCGGGCGACUUUGCUGCGGAUUCGGAGGCGG ....((((((((((((((((.(((((.(..(.(((((((((.((.(((((......))))).)).))))))))).)((((....)))).)))))).))))).)))))).)))))...... ( -55.10) >DroSim_CAF1 83153 120 + 1 GGGGCUGGGCUGCGGGGAGUAGUCCGGCACGGGGAAUCCCGAGGUGGCCACCACUAUGGUCUCCAUGGGAUUCUGCGAGGAUUGCUUCAGCGGGCGACUUUGCUGCGGGUUCGGAGGCGG ...((((((((((.....)))))))))).((.(((((((((.((.(((((......))))).)).))))))))).)).....(((((((((((....))..))).((....)))))))). ( -56.70) >DroEre_CAF1 35554 120 + 1 CGGGCUGGGCUGCGGGGAAUAGUCCGGUACGGGGAAUCCCGAGGUGGCCACCACAAUGGUUUCCAUGGGAUUCUGCGAGGCUUGUCUCAGCGGGCGGCUUUGCUGCGGAUUCGGAGGCGG ((..(((((((((((.(((..(((((.(..(.(((((((((.((..((((......))))..)).))))))))).)((((....))))).)))))...))).)))))).)))))...)). ( -53.50) >DroYak_CAF1 142313 120 + 1 GGGGCUGGGCUGCGGGGAAUAGUCCGGUACGGGGAAUCCCGAGGUGGCCACCACAAUGGUCUCCGUGGGAUUCUGCGACGAUGGCUUCAGCGGGCGACUUUGCUGCGGAUUCGGAGGAGG ....(((((((((((.(((..(((((((.((.(((((((((.((.(((((......))))).)).))))))))).))))((.....))..)))))...))).)))))).)))))...... ( -53.10) >consensus GGGGCUGGGCUGCGGGGAAUAGUCCGGCACGGGGAAUCCCGAGGUGGCCACCACAAUGGUCUCCAUGGGAUUCUGCGAGGAUUGCUUCAGCGGGCGACUUUGCUGCGGAUUCGGAGGCGG ....(((((((((((.(((..(((((....(.(((((((((.((.(((((......))))).)).))))))))).)((((....))))..)))))...))).)))))).)))))...... (-52.72 = -52.10 + -0.62)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:04:46 2006