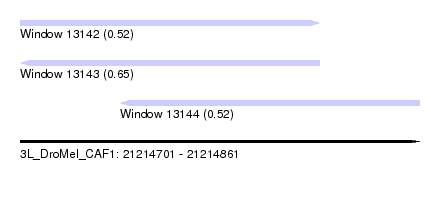
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,214,701 – 21,214,861 |
| Length | 160 |
| Max. P | 0.650847 |

| Location | 21,214,701 – 21,214,821 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.61 |
| Mean single sequence MFE | -34.87 |
| Consensus MFE | -31.49 |
| Energy contribution | -31.61 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.90 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21214701 120 + 23771897 GCAACUGCUGUUGACUAGGGCCGGUGUGUCCUCCACCUCGGCAUUCUUUUCAAUUACCUUGCCACUAACGGGUGAAUACACCUCGCUAGCCGCUUUAACGCUCUCCAGGGCCCCACAUUC .((((....))))....((((((((((((....((((..((((................)))).(....))))).))))))).........((......)).......)))))....... ( -33.59) >DroSim_CAF1 81763 120 + 1 GCAACUGCUGUUGACCAGGGCCGGUGUGUCCUCCACCUCGGCAUUCUUUUCGAUCACCUUGCCACUAACGGGUGAAUACACCUCGCUAGCAGCUUUCACGCUCUCCAGGGCCCCACAUUC ......(((((((.(.((((((((.(((.....))).)))))...........((((((..........)))))).....))).).)))))))......((((....))))......... ( -34.70) >DroEre_CAF1 34341 120 + 1 GCAACUACUGUUGACCAGGGCCGGUGUGUCCUCCACCUGGGCAUUCUUUUCGAUCACCUUGCCGCUAACGGGUGAAUACACCUCGCUAGCAGCUUUUACGCUCUCCAGGGCUCCACAUUC .((((....))))....((((((((((((((.......))))).((.....)).))))..((.((((.((((((....)))).)).)))).))...............))).))...... ( -35.80) >DroYak_CAF1 141023 120 + 1 GCAACUACUGUUGACCAGGGCCGGUGUGUCCUCCACCAGAGCAUUCUUUUCGAUCACCUUGCCGCUAACUGGUGAAUACACCUCGCUGGCCGCUUUCACGCUCUCCAGGGCUCCACAUUC .((((....))))....((((((((((((....(((((((((.....................)))..)))))).)))))))...((((..((......))...))))))).))...... ( -35.40) >consensus GCAACUACUGUUGACCAGGGCCGGUGUGUCCUCCACCUCGGCAUUCUUUUCGAUCACCUUGCCACUAACGGGUGAAUACACCUCGCUAGCAGCUUUCACGCUCUCCAGGGCCCCACAUUC .((((....))))....((((((((((((....((((((((((................)))).....)))))).)))))))..((.....))...............)))))....... (-31.49 = -31.61 + 0.13)



| Location | 21,214,701 – 21,214,821 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.61 |
| Mean single sequence MFE | -43.00 |
| Consensus MFE | -38.35 |
| Energy contribution | -38.85 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21214701 120 - 23771897 GAAUGUGGGGCCCUGGAGAGCGUUAAAGCGGCUAGCGAGGUGUAUUCACCCGUUAGUGGCAAGGUAAUUGAAAAGAAUGCCGAGGUGGAGGACACACCGGCCCUAGUCAACAGCAGUUGC .....(((((((...............((.(((((((.((((....))))))))))).))..((((.((.....)).))))..((((.......)))))))))))..((((....)))). ( -44.80) >DroSim_CAF1 81763 120 - 1 GAAUGUGGGGCCCUGGAGAGCGUGAAAGCUGCUAGCGAGGUGUAUUCACCCGUUAGUGGCAAGGUGAUCGAAAAGAAUGCCGAGGUGGAGGACACACCGGCCCUGGUCAACAGCAGUUGC ......((((((...............((..((((((.((((....))))))))))..))..((((.((.....)).))))..((((.......))))))))))...((((....)))). ( -45.80) >DroEre_CAF1 34341 120 - 1 GAAUGUGGAGCCCUGGAGAGCGUAAAAGCUGCUAGCGAGGUGUAUUCACCCGUUAGCGGCAAGGUGAUCGAAAAGAAUGCCCAGGUGGAGGACACACCGGCCCUGGUCAACAGUAGUUGC ......((.(((((((...((((....((((((((((.((((....)))))))))))))).......((.....))))))))))(((.....)))...)))))....((((....)))). ( -43.30) >DroYak_CAF1 141023 120 - 1 GAAUGUGGAGCCCUGGAGAGCGUGAAAGCGGCCAGCGAGGUGUAUUCACCAGUUAGCGGCAAGGUGAUCGAAAAGAAUGCUCUGGUGGAGGACACACCGGCCCUGGUCAACAGUAGUUGC ......((.(((....(((((((....((.((.(((..((((....)))).))).)).)).......((.....)))))))))((((.......)))))))))....((((....)))). ( -38.10) >consensus GAAUGUGGAGCCCUGGAGAGCGUGAAAGCGGCUAGCGAGGUGUAUUCACCCGUUAGCGGCAAGGUGAUCGAAAAGAAUGCCCAGGUGGAGGACACACCGGCCCUGGUCAACAGCAGUUGC .....(((((((...............((((((((((.((((....))))))))))))))..((((.((.....)).))))..((((.......)))))))))))..((((....)))). (-38.35 = -38.85 + 0.50)

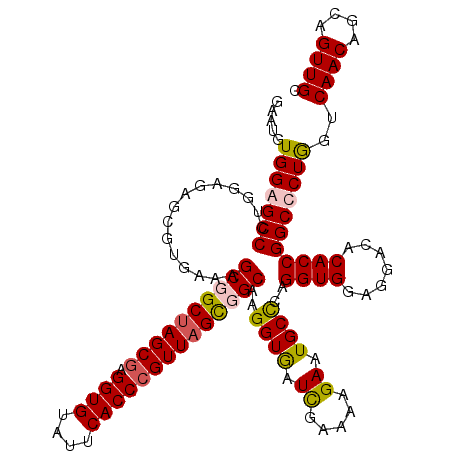
| Location | 21,214,741 – 21,214,861 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -40.75 |
| Consensus MFE | -36.47 |
| Energy contribution | -37.97 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.90 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21214741 120 - 23771897 CCAACUCCCAGAACCCGGCACGGAACUCAAGCAGGAUGACGAAUGUGGGGCCCUGGAGAGCGUUAAAGCGGCUAGCGAGGUGUAUUCACCCGUUAGUGGCAAGGUAAUUGAAAAGAAUGC ((..(((((((..(((.(((((..(..(.....)..)..))..))).)))..)))).))).......((.(((((((.((((....))))))))))).))..))................ ( -42.40) >DroSim_CAF1 81803 120 - 1 CCAGCUCCCAGAACCCGGCACGGAACUCAAGCAGGAUGACGAAUGUGGGGCCCUGGAGAGCGUGAAAGCUGCUAGCGAGGUGUAUUCACCCGUUAGUGGCAAGGUGAUCGAAAAGAAUGC ((.((((((((..(((.(((((..(..(.....)..)..))..))).)))..)))).))))......((..((((((.((((....))))))))))..))..))................ ( -47.70) >DroEre_CAF1 34381 120 - 1 UCAGCUGCCAGAACCCGGCACGGAACUCAAGCAGGAUGACGAAUGUGGAGCCCUGGAGAGCGUAAAAGCUGCUAGCGAGGUGUAUUCACCCGUUAGCGGCAAGGUGAUCGAAAAGAAUGC ((((((.((((..(.(.(((((..(..(.....)..)..))..))).).)..))))..)))......((((((((((.((((....))))))))))))))....)))............. ( -39.10) >DroYak_CAF1 141063 120 - 1 UCAGCUCCCAGAACCCGGCACGGAACUCAAACAGGAUGACGAAUGUGGAGCCCUGGAGAGCGUGAAAGCGGCCAGCGAGGUGUAUUCACCAGUUAGCGGCAAGGUGAUCGAAAAGAAUGC (((((((((((..(.(.(((((..(..(.....)..)..))..))).).)..)))).)))).)))..((.((.(((..((((....)))).))).)).)).................... ( -33.80) >consensus CCAGCUCCCAGAACCCGGCACGGAACUCAAGCAGGAUGACGAAUGUGGAGCCCUGGAGAGCGUGAAAGCGGCUAGCGAGGUGUAUUCACCCGUUAGCGGCAAGGUGAUCGAAAAGAAUGC ((.((((((((..(((.(((((..(..(.....)..)..))..))).)))..)))).))))......((((((((((.((((....))))))))))))))..))................ (-36.47 = -37.97 + 1.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:04:43 2006