| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,417,604 – 2,417,701 |
| Length | 97 |
| Max. P | 0.880472 |

| Location | 2,417,604 – 2,417,701 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 81.07 |
| Mean single sequence MFE | -33.38 |
| Consensus MFE | -21.38 |
| Energy contribution | -23.05 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
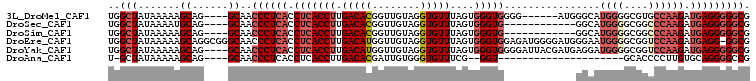
>3L_DroMel_CAF1 2417604 97 + 23771897 UGGCUAUAAAAAGCAG----GCAACCCUCACCUCACCUUGACACGGUUGUAGGUGUUUAGUGGGUGGGG------AUGGGCAUGGGGCGUGCCAAGAUGAGGGGGCG ..(((......)))..----((..((((((((((((((.(((((........)))))....))))))))------...((((((...))))))....)))))).)). ( -40.90) >DroSec_CAF1 50805 91 + 1 UGGCUAUAAAAUGCAG----GCAACCCUCACCUCACCUUGACACGGUUGUAGGUGUUUAGUGGGUG------------GGCAUGGGGCGGCCCAAGAUGAGGGGGCG ..((........))..----((..((((((.(((((((.(((((........)))))....)))))------------))..((((....))))...)))))).)). ( -33.40) >DroSim_CAF1 44737 91 + 1 UGGCUAUAAAAAGCAG----GCAACCCUCACCUCACCUUGACACGGUUGUAGGUGUUUAGUGGGUG------------GGCAUGGGGCGGCCCAAGAUGAGGGGGCG ..(((......)))..----((..((((((.(((((((.(((((........)))))....)))))------------))..((((....))))...)))))).)). ( -34.20) >DroEre_CAF1 44433 106 + 1 UGGCUAUAAAAAGCAGGCGGGCAACCCUCACCUCACCUUGACAUGGUUGUAGGUGUUUAGUGGGUGGAGAUGGGGAUGGGAAUGGGGCGGUCCAAGAUGAGG-GGCG ..((........((......))..(((((((..(((((.(((((........)))))....)))))..)....((((.(........).))))....)))))-))). ( -30.50) >DroYak_CAF1 45499 103 + 1 UGGCUAUAAAAAGCAG----GCAACCCUCACCUCACCUUGACAUGGUUGUAGGUGUUUAGUGGGUGGGGAUUACGAUGAGGAUGGGGCGGUCCAAGAUGAGGGGGCG ..(((......)))..----((..((((((((((((((.(((((........)))))....))))))))..........((((......))))....)))))).)). ( -35.10) >DroAna_CAF1 38543 79 + 1 U-GCUAUAAAAAGCAG----GCAACCCUCACCUCACCUUGACACGAUUGUGGGUGUUUCG--GGU---------------------GCACCCCUUGUGCAGGGGCCG (-(((......))))(----((..((((.(((((((((..(((....))))))))....)--)))---------------------((((.....))))))))))). ( -26.20) >consensus UGGCUAUAAAAAGCAG____GCAACCCUCACCUCACCUUGACACGGUUGUAGGUGUUUAGUGGGUG____________GGCAUGGGGCGGCCCAAGAUGAGGGGGCG ..(((.......((......))..((((((.(((((((.(((((........)))))....)))))................((((....)))))).))))))))). (-21.38 = -23.05 + 1.67)
| Location | 2,417,604 – 2,417,701 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 81.07 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -18.47 |
| Energy contribution | -20.13 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880472 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
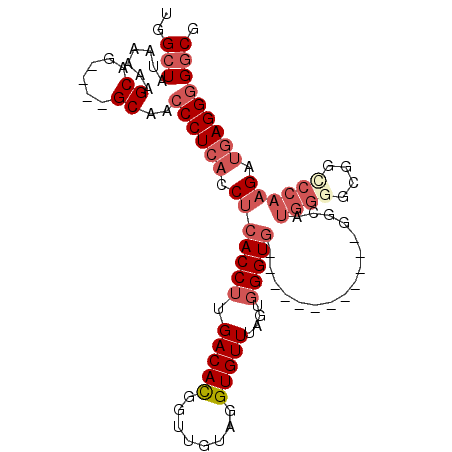
>3L_DroMel_CAF1 2417604 97 - 23771897 CGCCCCCUCAUCUUGGCACGCCCCAUGCCCAU------CCCCACCCACUAAACACCUACAACCGUGUCAAGGUGAGGUGAGGGUUGC----CUGCUUUUUAUAGCCA .(((((((((((((((((((............------........................)))))))))))))))...))))...----..(((......))).. ( -28.92) >DroSec_CAF1 50805 91 - 1 CGCCCCCUCAUCUUGGGCCGCCCCAUGCC------------CACCCACUAAACACCUACAACCGUGUCAAGGUGAGGUGAGGGUUGC----CUGCAUUUUAUAGCCA .((.((((((((((((((........)))------------)).........((((((((....)))..))))))))))))))..))----..((........)).. ( -30.00) >DroSim_CAF1 44737 91 - 1 CGCCCCCUCAUCUUGGGCCGCCCCAUGCC------------CACCCACUAAACACCUACAACCGUGUCAAGGUGAGGUGAGGGUUGC----CUGCUUUUUAUAGCCA .((.((((((((((((((........)))------------)).........((((((((....)))..))))))))))))))..))----..(((......))).. ( -31.60) >DroEre_CAF1 44433 106 - 1 CGCC-CCUCAUCUUGGACCGCCCCAUUCCCAUCCCCAUCUCCACCCACUAAACACCUACAACCAUGUCAAGGUGAGGUGAGGGUUGCCCGCCUGCUUUUUAUAGCCA .(((-(((((((((((......)))...........................((((((((....)))..))))))))))))))..))......(((......))).. ( -25.30) >DroYak_CAF1 45499 103 - 1 CGCCCCCUCAUCUUGGACCGCCCCAUCCUCAUCGUAAUCCCCACCCACUAAACACCUACAACCAUGUCAAGGUGAGGUGAGGGUUGC----CUGCUUUUUAUAGCCA .((.((((((((((((......)))...........................((((((((....)))..))))))))))))))..))----..(((......))).. ( -25.90) >DroAna_CAF1 38543 79 - 1 CGGCCCCUGCACAAGGGGUGC---------------------ACC--CGAAACACCCACAAUCGUGUCAAGGUGAGGUGAGGGUUGC----CUGCUUUUUAUAGC-A ..((((((.....))))))((---------------------(((--(....(((((((............))).)))).))).)))----..(((......)))-. ( -28.70) >consensus CGCCCCCUCAUCUUGGGCCGCCCCAUGCC____________CACCCACUAAACACCUACAACCGUGUCAAGGUGAGGUGAGGGUUGC____CUGCUUUUUAUAGCCA ....((((((((((((......)))...........................((((((((....)))..))))))))))))))..........(((......))).. (-18.47 = -20.13 + 1.67)

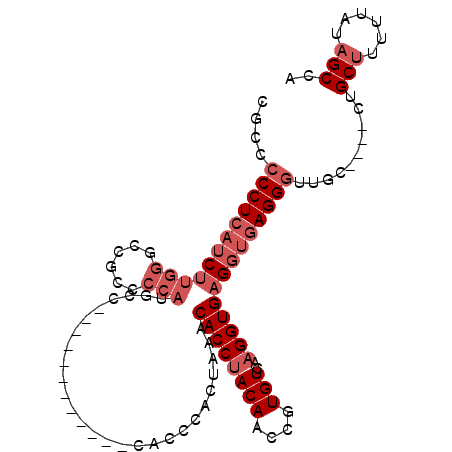
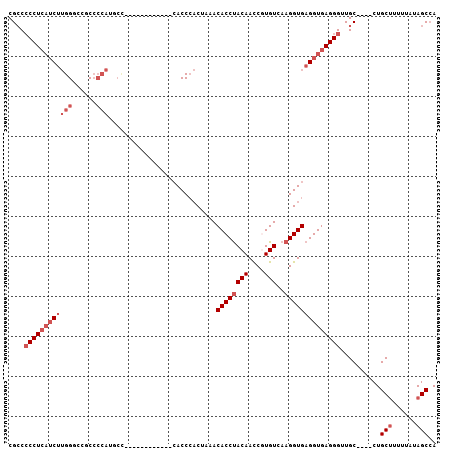
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:37 2006