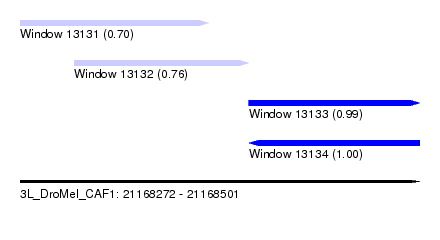
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,168,272 – 21,168,501 |
| Length | 229 |
| Max. P | 0.998504 |

| Location | 21,168,272 – 21,168,380 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.89 |
| Mean single sequence MFE | -29.68 |
| Consensus MFE | -15.32 |
| Energy contribution | -14.95 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21168272 108 + 23771897 GGCGACACAGUAACAUAGAAUCAAACUGCUC----------UCGACU-CAAUUGUAGUCAAUUUGAAAGCCAACUAUCAAAUGUCAGCUACAAGCAUCCGAGAUUCGUUC-GCCACGAUU ((((((.((((.............)))).((----------(((...-...(((((((..((((((.((....)).))))))....))))))).....)))))...).))-)))...... ( -27.92) >DroSec_CAF1 39246 108 + 1 GGCGACACAGUAACACAGAAUCAAACUGCUC----------UCGACU-CAAUUGUAGUCAAUUUGAAAGCCAACUAUCAAAUGUCAGCUACAAGCAUCCGAGAUUCGUUC-GCCACGAUU (((((.((.......(((.......))).((----------(((...-...(((((((..((((((.((....)).))))))....))))))).....)))))...))))-)))...... ( -28.10) >DroEre_CAF1 35344 108 + 1 GGCGACAUAGUAACACAGAAUCAAACUGCUC----------UCGACU-CAAUUGUAGUCAAUUUGAAAGCCAACUAUCAAAUGUCAGCUGCAAAGAUCCGAGAUUCGUUC-GCCACGAUU ((((((.........(((.......))).((----------(((..(-(..(((((((..((((((.((....)).))))))....))))))).))..)))))...).))-)))...... ( -27.60) >DroMoj_CAF1 66392 114 + 1 GGCGACAUAGUAACAUAGAA-----CAGCUCGACAUCGAGUCUGACUCCAAUUGUAGUCAAUUUGAAAGCAAACUAUCAAAUGUCAGCAACACACGG-AGAGAUCUGCUCUGUCGCGCUU .((((((.((((........-----(((((((....)))).))).((((...(((.((..((((((.((....)).))))))....)).)))...))-)).....)))).)))))).... ( -35.10) >consensus GGCGACACAGUAACACAGAAUCAAACUGCUC__________UCGACU_CAAUUGUAGUCAAUUUGAAAGCCAACUAUCAAAUGUCAGCUACAAACAUCCGAGAUUCGUUC_GCCACGAUU ((((((.....................................((((.(....).)))).((((((.((....)).)))))))))..............(((.....))).)))...... (-15.32 = -14.95 + -0.37)

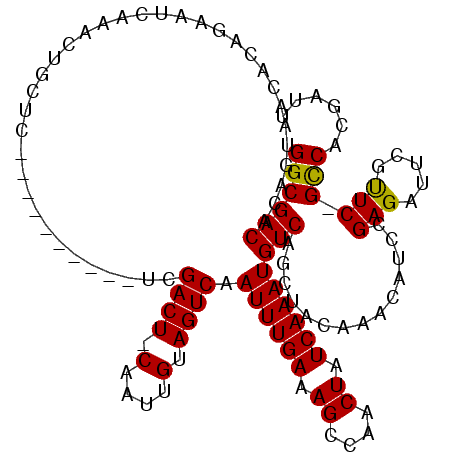
| Location | 21,168,303 – 21,168,403 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 77.83 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -16.15 |
| Energy contribution | -16.52 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21168303 100 + 23771897 -UCGACU-CAAUUGUAGUCAAUUUGAAAGCCAACUAUCAAAUGUCAGCUACAAGCAUCCGAGAUUCGUUC-GCCACGAUUCUUC----------UAGACGCCCAAAA-----UCGAGU -...(((-(..(((((((..((((((.((....)).))))))....)))))))((.((.((((.((((..-...)))).)))).----------..)).))......-----..)))) ( -24.10) >DroSec_CAF1 39277 100 + 1 -UCGACU-CAAUUGUAGUCAAUUUGAAAGCCAACUAUCAAAUGUCAGCUACAAGCAUCCGAGAUUCGUUC-GCCACGAUUCUUC----------UAGACGCCCAAAA-----UCGAGU -...(((-(..(((((((..((((((.((....)).))))))....)))))))((.((.((((.((((..-...)))).)))).----------..)).))......-----..)))) ( -24.10) >DroEre_CAF1 35375 100 + 1 -UCGACU-CAAUUGUAGUCAAUUUGAAAGCCAACUAUCAAAUGUCAGCUGCAAAGAUCCGAGAUUCGUUC-GCCACGAUUCUUC----------UAGACGCCCAAAA-----UCGAGU -...(((-(..(((((((..((((((.((....)).))))))....)))))))......((((.((((..-...)))).)))).----------.............-----..)))) ( -21.70) >DroMoj_CAF1 66427 115 + 1 UCUGACUCCAAUUGUAGUCAAUUUGAAAGCAAACUAUCAAAUGUCAGCAACACACGG-AGAGAUCUGCUCUGUCGCGCUUGUUGGAUGACGCUUGGAAUAGCCAAAAAAAAGACAA-- ..(((((.(....).)))))((((((.((....)).))))))((((.(((((..(((-(.(((.....))).)).))..)))))..))))..((((.....))))...........-- ( -26.90) >consensus _UCGACU_CAAUUGUAGUCAAUUUGAAAGCCAACUAUCAAAUGUCAGCUACAAACAUCCGAGAUUCGUUC_GCCACGAUUCUUC__________UAGACGCCCAAAA_____UCGAGU .((((......(((((((..((((((.((....)).))))))....)))))))......((((.((((......)))).)))).............................)))).. (-16.15 = -16.52 + 0.38)

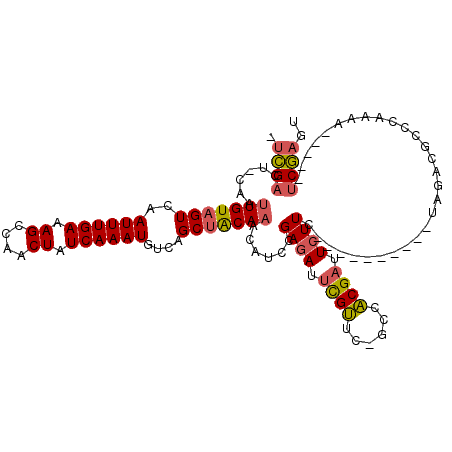
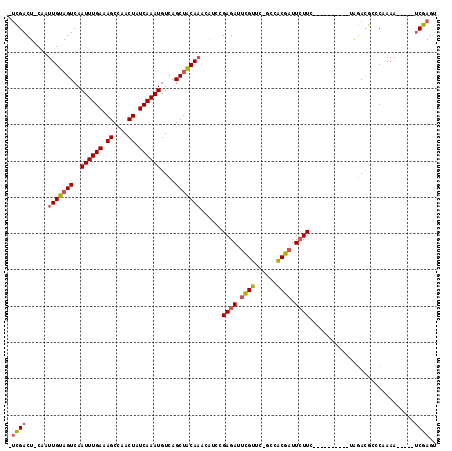
| Location | 21,168,403 – 21,168,501 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 79.51 |
| Mean single sequence MFE | -26.94 |
| Consensus MFE | -17.45 |
| Energy contribution | -17.23 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21168403 98 + 23771897 GCAAAACGCAACUGUUGCUGCCAAAAACUUCAUGUUGCCGCUGCGCCUGCGCAGUGACACCAGCGGUGACACCAGCAACAACAACGAAAACAACAGCC ((.....))...((((((((......(((...((.((.(((((((....))))))).)).))..))).....)))))))).................. ( -28.70) >DroSec_CAF1 39377 92 + 1 GCAAAACGCGACUGUUGCUGCCAAAAACUUCAUGUUGCCGCUGCGCCUGCGCAGUGACACCAGC------AACAACAAUUACAAAAACAACAACAGCC ((.....))..((((((.((............(((((((((((((....)))))))......))------)))).............)).)))))).. ( -24.61) >DroEre_CAF1 35475 79 + 1 GCGAAACGAAACUGUUGCUGCCAA-AACUUCAUGUUGCCGCUGCGCCUGCGCAGUGACACCGGC------AAC------AAC------AGCAAAAGCC ...........((((((.((((..-(((.....)))..(((((((....))))))).....)))------).)------)))------))........ ( -27.50) >consensus GCAAAACGCAACUGUUGCUGCCAAAAACUUCAUGUUGCCGCUGCGCCUGCGCAGUGACACCAGC______AACA_CAA_AACAA__A_AACAACAGCC ((..((((....))))((((.....(((.....)))..(((((((....)))))))....))))...............................)). (-17.45 = -17.23 + -0.22)

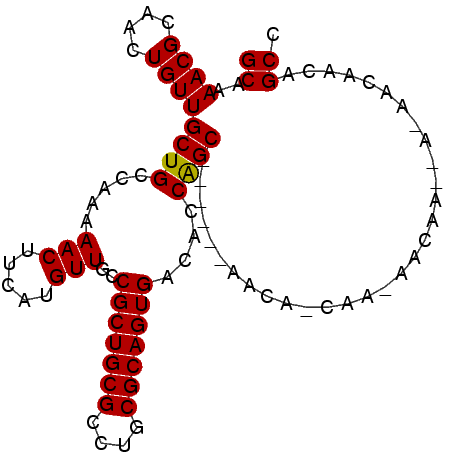

| Location | 21,168,403 – 21,168,501 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 79.51 |
| Mean single sequence MFE | -37.19 |
| Consensus MFE | -24.67 |
| Energy contribution | -24.57 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.41 |
| Structure conservation index | 0.66 |
| SVM decision value | 3.12 |
| SVM RNA-class probability | 0.998504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21168403 98 - 23771897 GGCUGUUGUUUUCGUUGUUGUUGCUGGUGUCACCGCUGGUGUCACUGCGCAGGCGCAGCGGCAACAUGAAGUUUUUGGCAGCAACAGUUGCGUUUUGC (((((((((((((((.(((((((((((((.((((...)))).))))(((....)))))))))))))))))((.....))))))))))))......... ( -40.00) >DroSec_CAF1 39377 92 - 1 GGCUGUUGUUGUUUUUGUAAUUGUUGUU------GCUGGUGUCACUGCGCAGGCGCAGCGGCAACAUGAAGUUUUUGGCAGCAACAGUCGCGUUUUGC (((((((((((((........(((((((------((((.((((........)))))))))))))))..........)))))))))))))......... ( -38.47) >DroEre_CAF1 35475 79 - 1 GGCUUUUGCU------GUU------GUU------GCCGGUGUCACUGCGCAGGCGCAGCGGCAACAUGAAGUU-UUGGCAGCAACAGUUUCGUUUCGC (((....(((------(((------(((------(((((.(((.(((((....))))).)))(((.....)))-))))))))))))))...))).... ( -33.10) >consensus GGCUGUUGUU_U__UUGUU_UUG_UGUU______GCUGGUGUCACUGCGCAGGCGCAGCGGCAACAUGAAGUUUUUGGCAGCAACAGUUGCGUUUUGC ((((((((((...........................(.((((.(((((....))))).)))).).....((.....))))))))))))......... (-24.67 = -24.57 + -0.11)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:04:33 2006