| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,166,997 – 21,167,157 |
| Length | 160 |
| Max. P | 0.949712 |

| Location | 21,166,997 – 21,167,117 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -39.10 |
| Consensus MFE | -36.14 |
| Energy contribution | -36.03 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
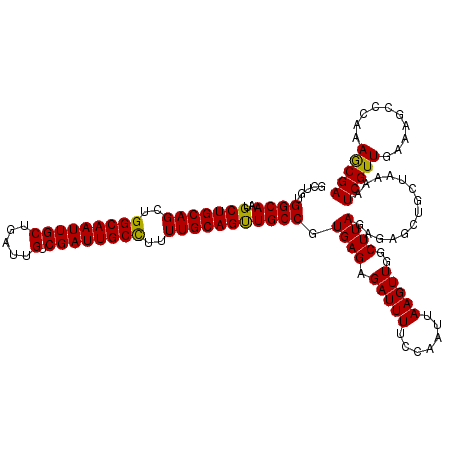
>3L_DroMel_CAF1 21166997 120 + 23771897 UGUCGAAUCGGAUGGGGUUAUUAUGGCUUAUGGGUUGUGGGCUGUGGCAAUGCUGCAGCUGGCAAUUGCUGAUUGUCGAUUGCUUUUUGCAGUUGCCGUGAGAGAUUUUCCAAUUAAGUU .........(((..(((((.(((((((.....((((((.(((((..((...))..))))).))))))..........((((((.....)))))))))))))..))))))))......... ( -39.40) >DroSec_CAF1 37957 120 + 1 UGUCGAAUCGGAUGGGGUUAUUAUGGCUUAUGGGUUGUGGGCUGUGGCAAUGCUGCAGCUGGCAAUUGCUGAUUGUCGAUUGCCUUUUGCAGUUGCCGUGAGAGAUUUUCCAAUUAAGUU ((..(((((.............(((((((((.....)))))))))((((..(((((((..(((((((((.....).))))))))..)))))))))))......)))))..))........ ( -39.70) >DroEre_CAF1 34081 113 + 1 UGUCGAAUCGGAUGGGGUUAUUAUGGCUUAUGGGUUG-------UGGCAAUGCUGCAGCUGGCAAUUGCUGUUGGUCGAUUGCCUUUUGCAGCUGCCGUGAGAGAUUUUCCAAUUAAGUU ((..(((((..(((((.(......).)))))...(..-------(((((..(((((((..(((((((((.....).))))))))..))))))))))))..)..)))))..))........ ( -38.20) >consensus UGUCGAAUCGGAUGGGGUUAUUAUGGCUUAUGGGUUGUGGGCUGUGGCAAUGCUGCAGCUGGCAAUUGCUGAUUGUCGAUUGCCUUUUGCAGUUGCCGUGAGAGAUUUUCCAAUUAAGUU .........(((..(((((.(((((((......(((((........)))))(((((((..(((((((((.....).))))))))..))))))).)))))))..))))))))......... (-36.14 = -36.03 + -0.11)


| Location | 21,166,997 – 21,167,117 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -24.58 |
| Consensus MFE | -21.99 |
| Energy contribution | -22.10 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21166997 120 - 23771897 AACUUAAUUGGAAAAUCUCUCACGGCAACUGCAAAAAGCAAUCGACAAUCAGCAAUUGCCAGCUGCAGCAUUGCCACAGCCCACAACCCAUAAGCCAUAAUAACCCCAUCCGAUUCGACA .....(((((((...........((((((((((....(((((.(........).)))))....)))))..)))))...((.............)).............)))))))..... ( -22.02) >DroSec_CAF1 37957 120 - 1 AACUUAAUUGGAAAAUCUCUCACGGCAACUGCAAAAGGCAAUCGACAAUCAGCAAUUGCCAGCUGCAGCAUUGCCACAGCCCACAACCCAUAAGCCAUAAUAACCCCAUCCGAUUCGACA .....(((((((...........((((((((((...((((((.(........).))))))...)))))..)))))...((.............)).............)))))))..... ( -25.62) >DroEre_CAF1 34081 113 - 1 AACUUAAUUGGAAAAUCUCUCACGGCAGCUGCAAAAGGCAAUCGACCAACAGCAAUUGCCAGCUGCAGCAUUGCCA-------CAACCCAUAAGCCAUAAUAACCCCAUCCGAUUCGACA .....(((((((...........((((((((((...((((((.(........).))))))...))))))..)))).-------.....(....)..............)))))))..... ( -26.10) >consensus AACUUAAUUGGAAAAUCUCUCACGGCAACUGCAAAAGGCAAUCGACAAUCAGCAAUUGCCAGCUGCAGCAUUGCCACAGCCCACAACCCAUAAGCCAUAAUAACCCCAUCCGAUUCGACA .....(((((((...........((((((((((...((((((.(........).))))))...)))))..))))).............(....)..............)))))))..... (-21.99 = -22.10 + 0.11)

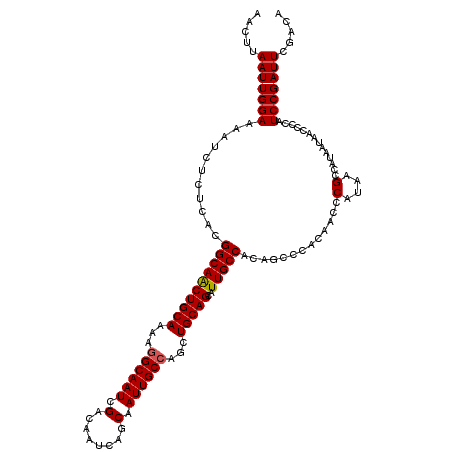
| Location | 21,167,037 – 21,167,157 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -37.57 |
| Consensus MFE | -33.93 |
| Energy contribution | -33.27 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21167037 120 + 23771897 GCUGUGGCAAUGCUGCAGCUGGCAAUUGCUGAUUGUCGAUUGCUUUUUGCAGUUGCCGUGAGAGAUUUUCCAAUUAAGUUGGCUUAGAGAGCUGCUAAAAUCGUUGAAAGCCCAAAGCGA (((.((((((((..(((((((((((((((.((..((.....))..)).))))))))).((((.(((((.......)))))..))))...))))))......))))).....))).))).. ( -36.90) >DroSec_CAF1 37997 120 + 1 GCUGUGGCAAUGCUGCAGCUGGCAAUUGCUGAUUGUCGAUUGCCUUUUGCAGUUGCCGUGAGAGAUUUUCCAAUUAAGUUGGCUUAGAGAGCUGCUAGAAUCGUUGAAAGCCCAAAGCGA (((..((((..(((((((..(((((((((.....).))))))))..)))))))))))......((((((.......(((.(((((...)))))))))))))).............))).. ( -37.01) >DroEre_CAF1 34118 115 + 1 ----UGGCAAUGCUGCAGCUGGCAAUUGCUGUUGGUCGAUUGCCUUUUGCAGCUGCCGUGAGAGAUUUUCCAAUUAAGUUGGCUUAGA-AGCUGAUAAAAUCGUUGAAAGCCUAAAACGA ----.((((..(((((((..(((((((((.....).))))))))..)))))))))))((...((..((((..(((..((..(((....-)))..))..)))....))))..))...)).. ( -38.80) >consensus GCUGUGGCAAUGCUGCAGCUGGCAAUUGCUGAUUGUCGAUUGCCUUUUGCAGUUGCCGUGAGAGAUUUUCCAAUUAAGUUGGCUUAGAGAGCUGCUAAAAUCGUUGAAAGCCCAAAGCGA .....((((..(((((((..(((((((((.....).))))))))..))))))))))).((((.(((((.......)))))..))))..............(((((..........))))) (-33.93 = -33.27 + -0.66)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:04:29 2006