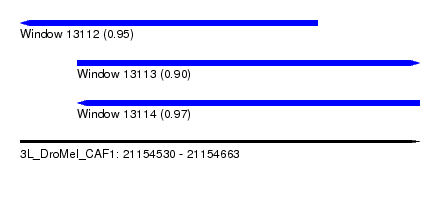
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,154,530 – 21,154,663 |
| Length | 133 |
| Max. P | 0.970699 |

| Location | 21,154,530 – 21,154,629 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 80.15 |
| Mean single sequence MFE | -36.86 |
| Consensus MFE | -24.99 |
| Energy contribution | -24.60 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21154530 99 - 23771897 CGUGGUGUUUGAAUACUCCAUGGAUUGCCGGCCCUCCAAGGGGCUGGCUGUUUUUCCGAGCUGUCACUAAAAUACAUUUAAGCAGCUGCAC------CAC-----CGAUU .(((((((............((((..(((((((((....)))))))))......))))((((((...((((.....)))).))))))))))------)))-----..... ( -38.20) >DroVir_CAF1 45531 104 - 1 CUUUGAGUUUGAAUACUCCAUGGAUUGCCGGCCCUCAAGGGGGCUGGCCGCUGUUGGUGGCAAGCACUAAAAUACAUUUAAACAGCUGCUGCCUGUGCGC-----CGAG- ....((((......))))..(((...(((((((((....)))))))))(((....((..(((.((...................)))))..))...))))-----))..- ( -38.51) >DroGri_CAF1 45528 109 - 1 CUUUGAGUUUGAAUACUCCAUGGAUUGCCGGCCCUCAAUGGGGCUGGCCGUUGUUGAGUGCAAGCACUAAAAUACAUUUAAACAGCUGCUGUUUGUGCCCUUUUCAGAU- .((((((((((..(((((.(..(...(((((((((....)))))))))..)..).)))))))))).............(((((((...)))))))........))))).- ( -34.90) >DroSec_CAF1 25576 99 - 1 CGUGGUGUUUGAAUACUCCAUGGAUUGCCGGCCCUCCAAGGGGCUGGCUGUUUUUCCGAGCUGUCACUAAAAUACAUUUAAGCAGCUGCAC------CAC-----CGAUU .(((((((............((((..(((((((((....)))))))))......))))((((((...((((.....)))).))))))))))------)))-----..... ( -38.20) >DroEre_CAF1 21941 99 - 1 AGUGGUGUUUGAAUACUCCAUGGAUUGCCGGCCCUCCAAGGGGCUGGCUGUUUUUCCGAGCUGUCACUAAAAUACAUUUAAGCAGCUGCAC------CAC-----CGAUU .(((((((............((((..(((((((((....)))))))))......))))((((((...((((.....)))).))))))))))------)))-----..... ( -38.10) >DroMoj_CAF1 49262 100 - 1 UUUAGAGUUUGAAUACUCCAUGGAUUGCCGGCCCUCAAGGGGGCUGGCCGCUGUUGGUUGCAAGCACUCAAAUACAUUUAAACAGCUGCUG----UGGGA-----GGUG- ...............((((((((...(((((((((....))))))))).((((((...((..............))....))))))..)))----).)))-----)...- ( -33.24) >consensus CGUGGAGUUUGAAUACUCCAUGGAUUGCCGGCCCUCAAAGGGGCUGGCCGUUGUUCCGAGCAAGCACUAAAAUACAUUUAAACAGCUGCAC______CAC_____CGAU_ ......((((((((............(((((((((....))))))))).((((....))))..............))))))))........................... (-24.99 = -24.60 + -0.39)

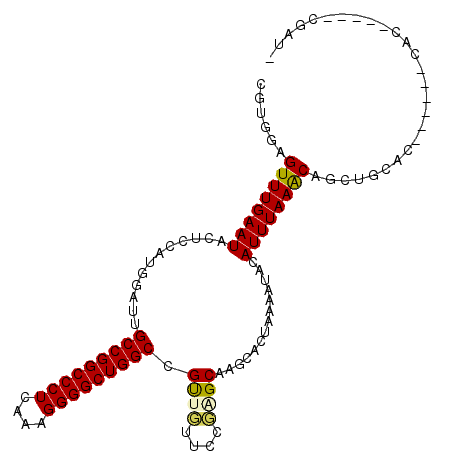

| Location | 21,154,549 – 21,154,663 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.89 |
| Mean single sequence MFE | -32.50 |
| Consensus MFE | -21.32 |
| Energy contribution | -21.16 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21154549 114 + 23771897 UAAAUGUAUUUUAGUGACAGCUCGGAAAAACAGCCAGCCCCUUGGAGGGCCGGCAAUCCAUGGAGUAUUCAAACACCACGGGUUUGACUCACAGCA------UCAAUGAGAACUCACAGA .............((((...((((((......(((.((((......)))).)))..))).(((((...((((((.(....)))))))))).))...------.....)))...))))... ( -31.50) >DroGri_CAF1 45557 101 + 1 UAAAUGUAUUUUAGUGCUUGCACUCAACAACGGCCAGCCCCAUUGAGGGCCGGCAAUCCAUGGAGUAUUCAAACUCAAAGUGUUUGAAUCACAACGAGGA---------G---------- ....(((..((.((((....)))).))..)))(((.((((......)))).)))..(((.((..((((((((((.......)))))))).))..)).)))---------.---------- ( -30.30) >DroSec_CAF1 25595 114 + 1 UAAAUGUAUUUUAGUGACAGCUCGGAAAAACAGCCAGCCCCUUGGAGGGCCGGCAAUCCAUGGAGUAUUCAAACACCACGGGUUUGACUCACAGCA------UCAACGAGAACUCACAGA .............((((...((((........(((.((((......)))).)))......(((((...((((((.(....)))))))))).))...------....))))...))))... ( -33.20) >DroEre_CAF1 21960 114 + 1 UAAAUGUAUUUUAGUGACAGCUCGGAAAAACAGCCAGCCCCUUGGAGGGCCGGCAAUCCAUGGAGUAUUCAAACACCACUGGUUUGACUCACAACA------UCAAGGAGAAGUCGCACA .............(((((..((((((......(((.((((......)))).)))..))).(((((...((((((.......))))))))).))...------.....)))..)))))... ( -33.20) >DroMoj_CAF1 49282 120 + 1 UAAAUGUAUUUGAGUGCUUGCAACCAACAGCGGCCAGCCCCCUUGAGGGCCGGCAAUCCAUGGAGUAUUCAAACUCUAAAUGUUUGAAUCACAACGAGCCAAUACAAGAGCAGCCACGGC ....(((.((((.(((((((...(((......(((.((((......)))).)))......))).((((((((((.......)))))))).))..)))))..)).)))).)))((....)) ( -34.30) >consensus UAAAUGUAUUUUAGUGACAGCUCGGAAAAACAGCCAGCCCCUUGGAGGGCCGGCAAUCCAUGGAGUAUUCAAACACCACGGGUUUGACUCACAACA______UCAA_GAGAACUCACAGA .............(((.............((.(((.((((......)))).)))..((....))))..((((((.......))))))..)))............................ (-21.32 = -21.16 + -0.16)

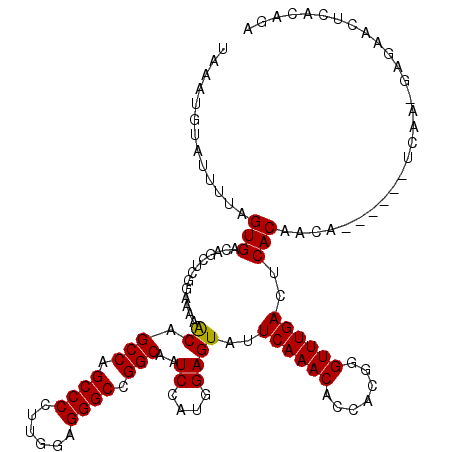
| Location | 21,154,549 – 21,154,663 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.89 |
| Mean single sequence MFE | -37.26 |
| Consensus MFE | -24.58 |
| Energy contribution | -24.78 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21154549 114 - 23771897 UCUGUGAGUUCUCAUUGA------UGCUGUGAGUCAAACCCGUGGUGUUUGAAUACUCCAUGGAUUGCCGGCCCUCCAAGGGGCUGGCUGUUUUUCCGAGCUGUCACUAAAAUACAUUUA ...((((...(((.((((------(.......)))))..((((((.((......)).))))))...(((((((((....))))))))).........)))...))))............. ( -37.90) >DroGri_CAF1 45557 101 - 1 ----------C---------UCCUCGUUGUGAUUCAAACACUUUGAGUUUGAAUACUCCAUGGAUUGCCGGCCCUCAAUGGGGCUGGCCGUUGUUGAGUGCAAGCACUAAAAUACAUUUA ----------.---------(((..(..(((.(((((((.......))))))))))..)..)))..(((((((((....))))))))).((.....((((....)))).....))..... ( -33.60) >DroSec_CAF1 25595 114 - 1 UCUGUGAGUUCUCGUUGA------UGCUGUGAGUCAAACCCGUGGUGUUUGAAUACUCCAUGGAUUGCCGGCCCUCCAAGGGGCUGGCUGUUUUUCCGAGCUGUCACUAAAAUACAUUUA ...((((...((((..((------.((............((((((.((......)).))))))...(((((((((....))))))))).)).))..))))...))))............. ( -39.70) >DroEre_CAF1 21960 114 - 1 UGUGCGACUUCUCCUUGA------UGUUGUGAGUCAAACCAGUGGUGUUUGAAUACUCCAUGGAUUGCCGGCCCUCCAAGGGGCUGGCUGUUUUUCCGAGCUGUCACUAAAAUACAUUUA .....(((..(((....(------((..(((..(((((((....).)))))).)))..)))(((..(((((((((....)))))))))......))))))..)))............... ( -35.60) >DroMoj_CAF1 49282 120 - 1 GCCGUGGCUGCUCUUGUAUUGGCUCGUUGUGAUUCAAACAUUUAGAGUUUGAAUACUCCAUGGAUUGCCGGCCCUCAAGGGGGCUGGCCGCUGUUGGUUGCAAGCACUCAAAUACAUUUA .((((((..(((........))).....(((.(((((((.......)))))))))).))))))...(((((((((....))))))))).(((((.....)).)))............... ( -39.50) >consensus UCUGUGACUUCUC_UUGA______UGUUGUGAGUCAAACCCGUGGUGUUUGAAUACUCCAUGGAUUGCCGGCCCUCCAAGGGGCUGGCUGUUUUUCCGAGCUGUCACUAAAAUACAUUUA ............................(((..((((((.......))))))..........(((.(((((((((....))))))))).)))............)))............. (-24.58 = -24.78 + 0.20)

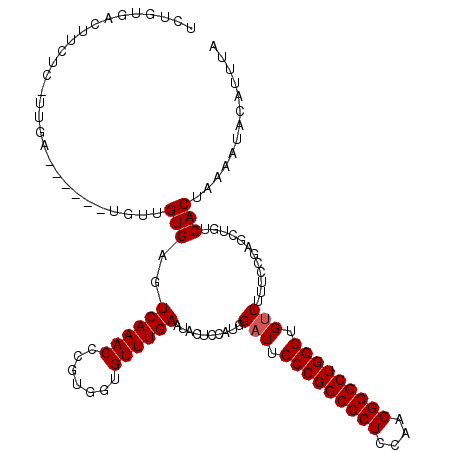
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:04:15 2006