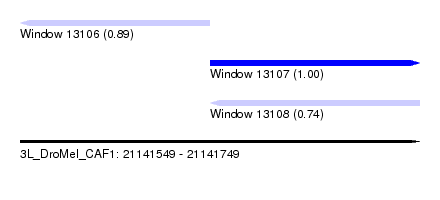
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,141,549 – 21,141,749 |
| Length | 200 |
| Max. P | 0.999711 |

| Location | 21,141,549 – 21,141,644 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 87.19 |
| Mean single sequence MFE | -22.70 |
| Consensus MFE | -16.46 |
| Energy contribution | -16.80 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21141549 95 - 23771897 AGAAAUAUGUUUUAAGUGCAACAUGGAGAUAUGUAACUAGCUUUAGUUCAGCACCAGUGCAAUCUAAUCUAACUGUUGCAAAUAUCAAUAUUUCU ((((((((....((..(((((((...((((.((.(((((....)))))))(((....)))......))))...)))))))..))...)))))))) ( -24.10) >DroSec_CAF1 12733 90 - 1 A-AAAUAUGUUUUAAGUGCAACAAGGAGAUAUGUAACUAGCUUUAGUUCAGCACCAGUGCA----AAUCUAAUUGUUGCAAAUAUCAAUAUUUCU .-((((((....((..((((((((..((((.((.(((((....)))))))(((....))).----.))))..))))))))..))...)))))).. ( -22.70) >DroEre_CAF1 8955 91 - 1 AGGAAUAUGCGUUAAGUGCAACAAGGAGAUAUGUAACAAGCUUUAAAUAGGCACCAGUGCA----AGUCGAACUUUUGCAAAUAUCAAUAUUUCU ((((((((....((..(((((.(((..(((.((((....((((.....)))).....))))----.)))...))))))))..))...)))))))) ( -21.30) >consensus AGAAAUAUGUUUUAAGUGCAACAAGGAGAUAUGUAACUAGCUUUAGUUCAGCACCAGUGCA____AAUCUAACUGUUGCAAAUAUCAAUAUUUCU ((((((((....((..(((((((...((((.((((....(((.......))).....)))).....))))...)))))))..))...)))))))) (-16.46 = -16.80 + 0.34)



| Location | 21,141,644 – 21,141,749 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.94 |
| Mean single sequence MFE | -32.53 |
| Consensus MFE | -18.99 |
| Energy contribution | -18.45 |
| Covariance contribution | -0.54 |
| Combinations/Pair | 1.20 |
| Mean z-score | -4.52 |
| Structure conservation index | 0.58 |
| SVM decision value | 3.93 |
| SVM RNA-class probability | 0.999711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21141644 105 + 23771897 AGUCUCCUAAGAGUCUGUCGUUGCUCCUAUUAUUUAGUUUUUAACUACGCA----------AGCUUU----CUUAUUAACUAAAAACAUGAGCUUCGG-CAUUCUUUUAGGGAAUUAGCA (((..((((((((..(((((..((((......(((((((..(((...(...----------.)....----.)))..))))))).....))))..)))-))..))))))))..))).... ( -31.40) >DroSec_CAF1 12823 106 + 1 AGUUUCCUAAAAGUCUGUCGUUGCUCCUAUUAUUUAGUUUUUAACUACGCA----------AGCUUU----CUUAUUAACUAAAAAUAUGAGCUUCGGACAUUCUUUUAGGGAAUUAGUA (((((((((((((..(((((..((((.((((.(((((((..(((...(...----------.)....----.)))..))))))))))).))))..).))))..))))))))))))).... ( -33.80) >DroEre_CAF1 9046 114 + 1 AGU-UCCCAAAAGAUCGUCGUUGCUC---UUUUUUAGUU-AUAACUACGCAGGAUUAGGUGAGCUUUCAAUCUGAAAGGCUAAAAAUCUGAGUUUCGA-CACUCUUUUGUUGAAUUAGUA (((-((.(((((((..((((..((((---.((((((((.-.......(((........)))..(((((.....)))))))))))))...))))..)))-)..)))))))..))))).... ( -32.40) >consensus AGU_UCCUAAAAGUCUGUCGUUGCUCCUAUUAUUUAGUUUUUAACUACGCA__________AGCUUU____CUUAUUAACUAAAAAUAUGAGCUUCGG_CAUUCUUUUAGGGAAUUAGUA (((.(((((((((..(((((..((((......(((((((.........((............)).............))))))).....))))..))).))..))))))))).))).... (-18.99 = -18.45 + -0.54)


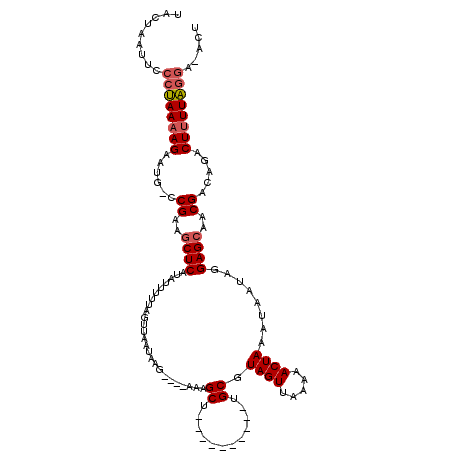
| Location | 21,141,644 – 21,141,749 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.94 |
| Mean single sequence MFE | -24.43 |
| Consensus MFE | -12.34 |
| Energy contribution | -13.23 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21141644 105 - 23771897 UGCUAAUUCCCUAAAAGAAUG-CCGAAGCUCAUGUUUUUAGUUAAUAAG----AAAGCU----------UGCGUAGUUAAAAACUAAAUAAUAGGAGCAACGACAGACUCUUAGGAGACU .......(((((((.((..((-.((..((((.(((((((((((..(((.----...(..----------..)....)))..))))))).)))).))))..)).))..)).))))).)).. ( -22.60) >DroSec_CAF1 12823 106 - 1 UACUAAUUCCCUAAAAGAAUGUCCGAAGCUCAUAUUUUUAGUUAAUAAG----AAAGCU----------UGCGUAGUUAAAAACUAAAUAAUAGGAGCAACGACAGACUUUUAGGAAACU .........((((((((..((((.(..((((.(((((((((((..(((.----...(..----------..)....)))..))))))).)))).))))..)))))..))))))))..... ( -26.60) >DroEre_CAF1 9046 114 - 1 UACUAAUUCAACAAAAGAGUG-UCGAAACUCAGAUUUUUAGCCUUUCAGAUUGAAAGCUCACCUAAUCCUGCGUAGUUAU-AACUAAAAAA---GAGCAACGACGAUCUUUUGGGA-ACU ......(((..(((((((.((-(((...(((.((((...(((.((((.....))))))).....)))).....((((...-.)))).....---)))...))))).))))))).))-).. ( -24.10) >consensus UACUAAUUCCCUAAAAGAAUG_CCGAAGCUCAUAUUUUUAGUUAAUAAG____AAAGCU__________UGCGUAGUUAAAAACUAAAUAAUAGGAGCAACGACAGACUUUUAGGA_ACU .........((((((((......((..((((.........................((............)).((((.....))))........))))..)).....))))))))..... (-12.34 = -13.23 + 0.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:04:09 2006