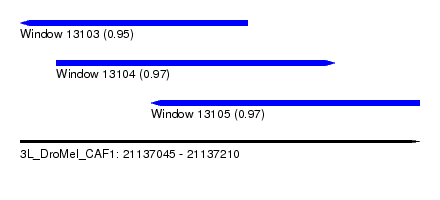
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,137,045 – 21,137,210 |
| Length | 165 |
| Max. P | 0.973678 |

| Location | 21,137,045 – 21,137,139 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 81.98 |
| Mean single sequence MFE | -34.58 |
| Consensus MFE | -23.16 |
| Energy contribution | -23.97 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.945127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21137045 94 - 23771897 GCGCCUGCGCAGCUGCAACAGCUGAUGUUGCUGUUGUUGCUGUUGUUUUGCUGGACAAAAGUACAGCAGCAUGUCGCCCACUAACGUGCAAAUG ((((.((((((((.((((((((.......)))))))).))))).....(((((.((....)).)))))))).).))).(((....)))...... ( -38.20) >DroSec_CAF1 8249 94 - 1 GCGCCUGCGCAGCUGUAGCAGCUGAUGCUGCUGUUGUUGCUGUUGUUUUGCUGGCCAAAAGUGCAGCAGCAUGUCGCCCACUAACGUGCAAAUG ((((.((((((((..(((((((....)))))))..)))))((((((...(((.......)))))))))))).).))).(((....)))...... ( -36.80) >DroEre_CAF1 4789 85 - 1 GCGCCUGCGCAGCUGCAGCAGCUGAUGCUGC---------UGUUGUUUUGCUGGCCAAAAGCGCAGCAGCAUGUCGCCCACUUACGUGCAAGUG ((((.(((..(((.((((((((....)))))---------))).))).(((((((.....)).)))))))).).))).(((((......))))) ( -34.90) >DroYak_CAF1 8329 76 - 1 GCA---------CUGCAGCAGCUGAUGUUGC---------UGUUGUUUUGCUGGCCGAAAGUGCAGCAGCAUGUCGCCCACCUACGUGCAACUG ..(---------(.((((((((....)))))---------))).))..(((((..(....)..)))))((((((.(.....).))))))..... ( -28.40) >consensus GCGCCUGCGCAGCUGCAGCAGCUGAUGCUGC_________UGUUGUUUUGCUGGCCAAAAGUGCAGCAGCAUGUCGCCCACUAACGUGCAAAUG ((((....(((((.((((((.....)))))).)))))...(((((((.((((.......)))).)))))))..............))))..... (-23.16 = -23.97 + 0.81)

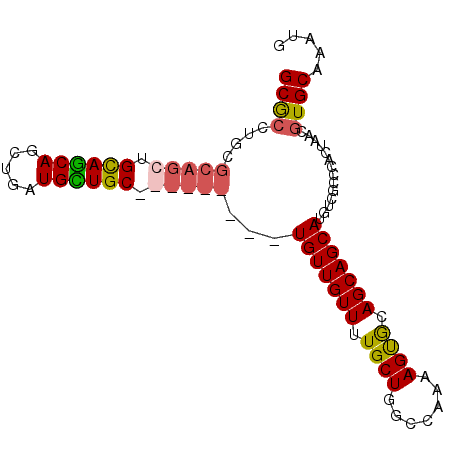
| Location | 21,137,060 – 21,137,175 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 73.93 |
| Mean single sequence MFE | -35.66 |
| Consensus MFE | -18.15 |
| Energy contribution | -21.94 |
| Covariance contribution | 3.79 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21137060 115 + 23771897 GGGCGACAUGCUG-CUGUACUUUUGUCCAGCAAAACAACAGCAACAACAGCAACAUCAGCUGUUGCAGCUGCGCAGGCGCCAAAUGCGUAUGCUAAUGUUGGCGGUAAGUUCAUAA ((((....(((((-(((.((....)).))))..((((.((((..(((((((.......)))))))..)))).(((.((((.....)))).)))...))))))))....)))).... ( -41.20) >DroSec_CAF1 8264 115 + 1 GGGCGACAUGCUG-CUGCACUUUUGGCCAGCAAAACAACAGCAACAACAGCAGCAUCAGCUGCUACAGCUGCGCAGGCGCCAAAUGCGCAUGCUAAUGUUGGCGGUAAGUUCAAAA ((((....(((((-(((..(....)..)))))...((((((((.(...((((((....))))))...).)))(((.((((.....)))).)))...))))))))....)))).... ( -41.10) >DroEre_CAF1 4804 106 + 1 GGGCGACAUGCUG-CUGCGCUUUUGGCCAGCAAAACAACA---------GCAGCAUCAGCUGCUGCAGCUGCGCAGGCGCCAAAUGCGCAUGCUAAUGUUGGCGGUAAGUUCAAAG .(.((((((...(-((((((.((((((...........((---------(((((....)))))))..(((.....))))))))).))))).))..)))))).)............. ( -44.30) >DroYak_CAF1 8344 97 + 1 GGGCGACAUGCUG-CUGCACUUUCGGCCAGCAAAACAACA---------GCAACAUCAGCUGCUGCAG---------UGCCAAAUGGCUAUGCUAAUGUUGGCGGUAAGUUCAAAA .((((......))-))(.((((.(.(((((((......((---------((.......))))..(((.---------.(((....)))..)))...))))))).).)))).).... ( -30.60) >DroAna_CAF1 27063 88 + 1 GGGCGG----CUGCCUGCACUUUUAGC---------AACAACAACAACAGCA---CUA------------GCGAGGGCGCCAAAUUCGCAUGCAAAUGGAAGCGGUAAGUUCAAAA ((((((----(.((((((.......))---------.............((.---...------------))..)))))))....((((.(.(....).).))))...)))).... ( -21.10) >consensus GGGCGACAUGCUG_CUGCACUUUUGGCCAGCAAAACAACA_CAACAACAGCAACAUCAGCUGCUGCAGCUGCGCAGGCGCCAAAUGCGCAUGCUAAUGUUGGCGGUAAGUUCAAAA .(((.....)))...((.((((.(.(((((((.................((((((.....))))))(((((((((.........)))))).)))..))))))).).)))).))... (-18.15 = -21.94 + 3.79)

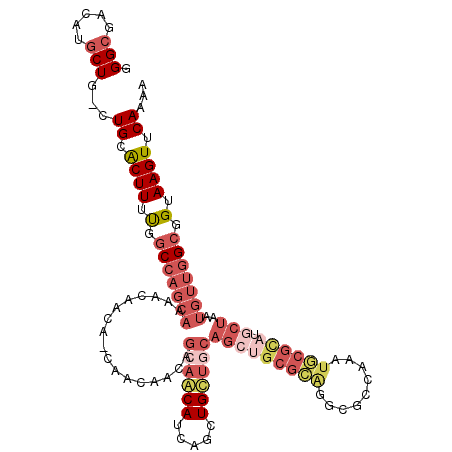
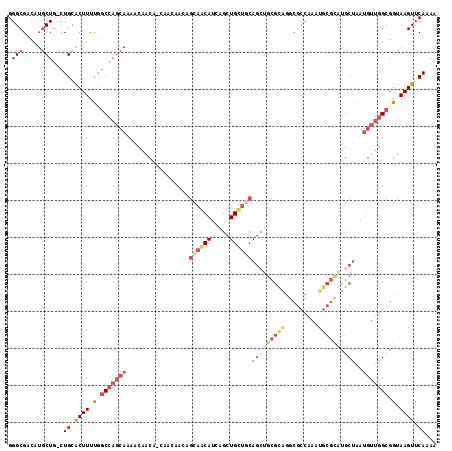
| Location | 21,137,099 – 21,137,210 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 73.98 |
| Mean single sequence MFE | -32.94 |
| Consensus MFE | -17.15 |
| Energy contribution | -19.46 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21137099 111 - 23771897 UCACCGAGUGCACGCGCUGUUUGCAUAAAUUAAGCUUAUGAACUUACCGCCAACAUUAGCAUACGCAUUUGGCGCCUGCGCAGCUGCAACAGCUGAUGUUGCUGUUGUUGC .......(((((.((((((..(((......((((........))))..((........))....)))..)))))).))))).((.((((((((.......)))))))).)) ( -38.20) >DroSec_CAF1 8303 111 - 1 UCACCGAGUGCACGCGCUAUUUGUAUAAAUUGAGCUUUUGAACUUACCGCCAACAUUAGCAUGCGCAUUUGGCGCCUGCGCAGCUGUAGCAGCUGAUGCUGCUGUUGUUGC ...((((((((.((.((((..(((......((((........))))......))).)))).)).)))))))).......(((((..(((((((....)))))))..))))) ( -38.10) >DroEre_CAF1 4843 102 - 1 UUACCAAGUGCACGCGCUAUUUGCACAGAUACAGCCUUUGAACUUACCGCCAACAUUAGCAUGCGCAUUUGGCGCCUGCGCAGCUGCAGCAGCUGAUGCUGC--------- .......(((((.((((((..(((.((((.......))))........((........))....)))..)))))).)))))....((((((.....))))))--------- ( -34.20) >DroYak_CAF1 8383 93 - 1 UUACCGAGUGCCCGCGCUAUUUGCAUAAAUGGAGCUUUUGAACUUACCGCCAACAUUAGCAUAGCCAUUUGGCA---------CUGCAGCAGCUGAUGUUGC--------- ......((((((...((.....))..((((((.(((..((.............))..)))....))))))))))---------))((((((.....))))))--------- ( -25.52) >DroAna_CAF1 27090 95 - 1 UCAGCAAUUGCACGCGCAAUUUGCAUAAUUAAAG-UUUUGAACUUACCGCUUCCAUUUGCAUGCGAAUUUGGCGCCCUCGC------------UAG---UGCUGUUGUUGU .(((((((.((((....(((((((((.....(((-(.....))))...((........)))))))))))(((((....)))------------)))---))).))))))). ( -28.70) >consensus UCACCGAGUGCACGCGCUAUUUGCAUAAAUAAAGCUUUUGAACUUACCGCCAACAUUAGCAUGCGCAUUUGGCGCCUGCGCAGCUGCAGCAGCUGAUGCUGCUGUUGUUG_ .......(((((.((((((..(((.((....(((........)))...((........)).)).)))..)))))).)))))....((((((.....))))))......... (-17.15 = -19.46 + 2.31)

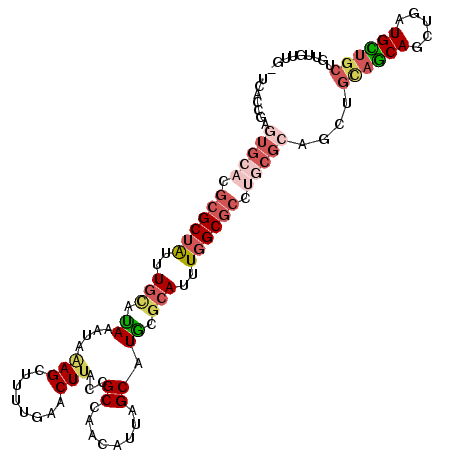
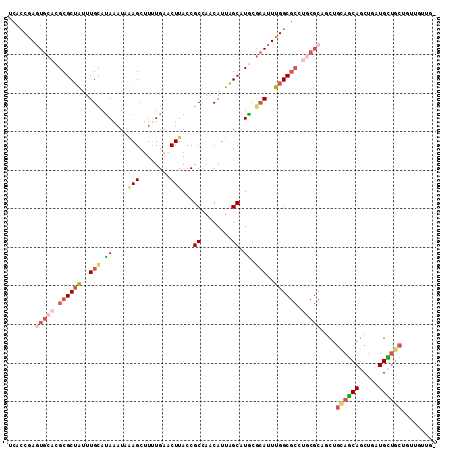
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:04:07 2006