| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,130,996 – 21,131,098 |
| Length | 102 |
| Max. P | 0.822593 |

| Location | 21,130,996 – 21,131,098 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.17 |
| Mean single sequence MFE | -29.26 |
| Consensus MFE | -18.17 |
| Energy contribution | -18.17 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.822593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
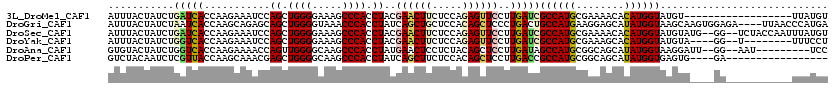
>3L_DroMel_CAF1 21130996 102 + 23771897 AUUUACUAUCUGAUCACCAAGAAAUCCAGCUGGGGAAAGCCCACCUACGAACUUCUCCAGAGUUCCUUGAUCGCCAUGCGAAAACACAUGGUAUGU------------------UUAUGU ...........(((((...........((.((((.....)))).))..((((((.....))))))..)))))((((((.(....).))))))....------------------...... ( -23.40) >DroGri_CAF1 22346 116 + 1 AUUUACUAUCUAAUCACCAAGCAGAGCAGCUGGGGUAAACCCACCUAUCAGCUGCUCCACAGCUCCCUGACUGCCAUGAAGGAGCAUAUGGUAAGCAAGUGGAGA----UUAACCCAUGA ..........(((((.(((.((.(((((((((((((......))))..)))))))))....(((((..............))))).........))...))).))----)))........ ( -36.64) >DroSec_CAF1 2203 116 + 1 AUUUACUAUCUGAUCACCAAGAAAUCCAGCUGGGGAAAGCCCACCUACGAACUUCUCCAGAGUUCCUUGAUCGCCAUGCGAAAACACAUGGUAUGUAUG--GG--UCUACCAAUUUAUGU ...(((.....(((((...........((.((((.....)))).))..((((((.....))))))..)))))((((((.(....).))))))..)))((--(.--....)))........ ( -26.60) >DroYak_CAF1 2104 106 + 1 AUUUACUAUCUGGUCACCAAGAAAUCCAGCUGGGGAAAGCCCACCUACGAACUUCUCCAGAGUUCCUUGAUCGCCAUGCGAAAGCACAUGGUAUGUA----GG--U--------UUUCCU .....(((.((((............)))).)))((((((((.((....((((((.....)))))).......(((((((....)))..))))..)).----))--)--------))))). ( -32.80) >DroAna_CAF1 21136 107 + 1 GUGUACUAUCUGGUCACCAAGAAAACCAGUUGGGGCAAGCCCACCUAUGAACUCCUCUACAGCUCCUUGAUAGCCAUGCGGCAGCAUAUGGUAAGGAUU--GG--AAU---------UCC (((.(((....))))))...(((..((((((((((....)))......((.....))..))))(((((....(((((((....)))..))))))))).)--))--..)---------)). ( -24.30) >DroPer_CAF1 10879 99 + 1 GUCUACAAUCUCGUUACCAAGCAAACGAGCUGGGGCAAGCCCACCUAUCAGCUUCUCCACAGCUCCUUGACCGCCAUGCGGCAGCAUAUGGUGAGUG----GA----------------- .(((((.......((((((.((....(((((((((.((((..........)))).))).))))))..((.((((...))))))))...)))))))))----))----------------- ( -31.81) >consensus AUUUACUAUCUGAUCACCAAGAAAACCAGCUGGGGAAAGCCCACCUACGAACUUCUCCACAGCUCCUUGAUCGCCAUGCGAAAGCACAUGGUAAGUA____GG___________UUAUGU ...........(((((...........((.((((.....)))).))..((((((.....))))))..)))))((((((........))))))............................ (-18.17 = -18.17 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:04:02 2006