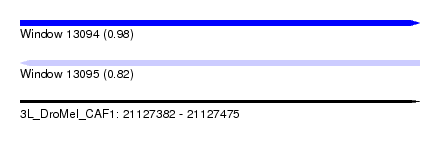
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,127,382 – 21,127,475 |
| Length | 93 |
| Max. P | 0.984528 |

| Location | 21,127,382 – 21,127,475 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 95.81 |
| Mean single sequence MFE | -29.18 |
| Consensus MFE | -27.52 |
| Energy contribution | -27.04 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21127382 93 + 23771897 UCGAUGGGCAACCUGCCCACAUGCGCGAGCGAGCUUUUCCUUU------UCGCCAUUUUCGAGGGCGGAAAAUGCGCUGCACUUUGCUCUUCUCCUCUC ..(.(((((.....))))))..((....))((((......(((------(((((.........)))))))).(((...)))....)))).......... ( -28.10) >DroSec_CAF1 178094 93 + 1 UCGAUGGGCAACCUGCCCACAUGCGCGAGCGAGCUUUUCCUUU------UCGCCAUUUUCGAGGGCGGAAAAUGCGCUGCACUUUGCUCUCCCCCUCUC ..(.(((((.....))))))(((.(((((.(((......))))------)))))))....(((((.(((....(((........)))..)))))))).. ( -29.70) >DroSim_CAF1 89786 93 + 1 UCGAUGGGCAACCUGCCCACAUGCGCGAGCGAGCUUUUCCUUU------UCGCCAUUUUCGAGGGCGGAAAAUGCGCUGCACUUUGCUCUCCCCCUCUC ..(.(((((.....))))))(((.(((((.(((......))))------)))))))....(((((.(((....(((........)))..)))))))).. ( -29.70) >DroEre_CAF1 100492 93 + 1 UCGAUGGGCAACCUGCCCACAUGCGCGAGCGAGCUUUUCCUUU------UCGCCAUUUUUGAGGGCGGAAAAUGCGCUGCACUUUGCUCUCCCCCUCUC ..(.(((((.....))))))(((.(((((.(((......))))------)))))))....(((((.(((....(((........)))..)))))))).. ( -29.40) >DroYak_CAF1 71439 99 + 1 UCGAUGGGCAACCUGCCCACAUGCGCGAGCGAGCUUUUCCUUUUCAAUUUCGCCAUUUUCGAGGGCGGAAAAUGCACUGCACUUUGCUCUCCCCCUCUC .((((((((.....)))))..(((....)))..................)))........(((((.(((....(((........)))..)))))))).. ( -29.00) >consensus UCGAUGGGCAACCUGCCCACAUGCGCGAGCGAGCUUUUCCUUU______UCGCCAUUUUCGAGGGCGGAAAAUGCGCUGCACUUUGCUCUCCCCCUCUC .((((((((.....)))))..(((....)))..................)))........(((((.(((....(((........)))..)))))))).. (-27.52 = -27.04 + -0.48)

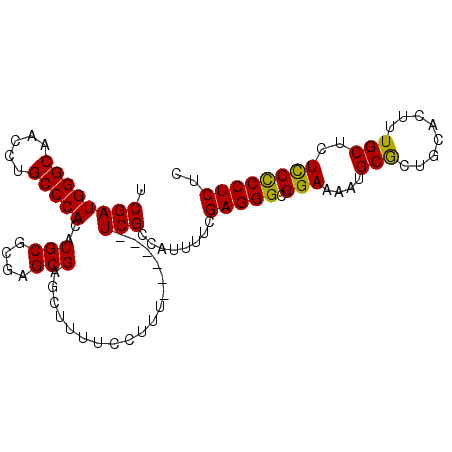
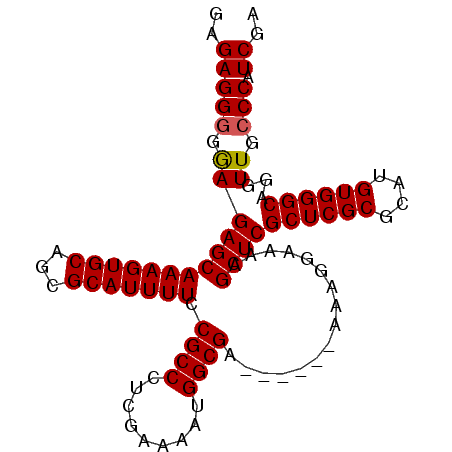
| Location | 21,127,382 – 21,127,475 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 95.81 |
| Mean single sequence MFE | -31.48 |
| Consensus MFE | -29.98 |
| Energy contribution | -30.02 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21127382 93 - 23771897 GAGAGGAGAAGAGCAAAGUGCAGCGCAUUUUCCGCCCUCGAAAAUGGCGA------AAAGGAAAAGCUCGCUCGCGCAUGUGGGCAGGUUGCCCAUCGA .................((((((((..((((((.(((........)).).------...))))))...)))).))))..((((((.....))))))... ( -29.50) >DroSec_CAF1 178094 93 - 1 GAGAGGGGGAGAGCAAAGUGCAGCGCAUUUUCCGCCCUCGAAAAUGGCGA------AAAGGAAAAGCUCGCUCGCGCAUGUGGGCAGGUUGCCCAUCGA ..((((((((((.....((((...))))))))).)))))........(((------...((...(((..((((((....))))))..))).))..))). ( -32.10) >DroSim_CAF1 89786 93 - 1 GAGAGGGGGAGAGCAAAGUGCAGCGCAUUUUCCGCCCUCGAAAAUGGCGA------AAAGGAAAAGCUCGCUCGCGCAUGUGGGCAGGUUGCCCAUCGA ..((((((((((.....((((...))))))))).)))))........(((------...((...(((..((((((....))))))..))).))..))). ( -32.10) >DroEre_CAF1 100492 93 - 1 GAGAGGGGGAGAGCAAAGUGCAGCGCAUUUUCCGCCCUCAAAAAUGGCGA------AAAGGAAAAGCUCGCUCGCGCAUGUGGGCAGGUUGCCCAUCGA ..((((((((((.....((((...))))))))).)))))........(((------...((...(((..((((((....))))))..))).))..))). ( -32.10) >DroYak_CAF1 71439 99 - 1 GAGAGGGGGAGAGCAAAGUGCAGUGCAUUUUCCGCCCUCGAAAAUGGCGAAAUUGAAAAGGAAAAGCUCGCUCGCGCAUGUGGGCAGGUUGCCCAUCGA ..(((((((((((((........)))..))))).))))).............((((...((...(((..((((((....))))))..))).))..)))) ( -31.60) >consensus GAGAGGGGGAGAGCAAAGUGCAGCGCAUUUUCCGCCCUCGAAAAUGGCGA______AAAGGAAAAGCUCGCUCGCGCAUGUGGGCAGGUUGCCCAUCGA ..(((((.(((((((((((((...))))))).((((.........))))................))))((((((....))))))...)).))).)).. (-29.98 = -30.02 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:03:57 2006